4ZGD
 
 | | Mutant R157A of Fe-Type Nitrile Hydratase from Comamonas testosteroni Ni1 | | Descriptor: | FE (III) ION, Nitrile hydratase alpha subunit, Nitrile hydratase beta subunit | | Authors: | Wu, R, Martinez, S, Holz, R, Liu, D. | | Deposit date: | 2015-04-22 | | Release date: | 2015-07-01 | | Last modified: | 2015-07-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Analyzing the catalytic role of active site residues in the Fe-type nitrile hydratase from Comamonas testosteroni Ni1.
J.Biol.Inorg.Chem., 20, 2015
|
|
7KEC
 
 | |
8BUE
 
 | | Structure of DDB1 bound to Z11-engaged CDK12-cyclin K | | Descriptor: | Cyclin-K, Cyclin-dependent kinase 12, DNA damage-binding protein 1, ... | | Authors: | Kozicka, Z, Kempf, G, Petzold, G, Thoma, N.H. | | Deposit date: | 2022-11-30 | | Release date: | 2023-09-13 | | Last modified: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
7L0U
 
 | |
7KKL
 
 | | SARS-CoV-2 Spike in complex with neutralizing nanobody mNb6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Schoof, M.S, Faust, B.F, Saunders, R.A, Sangwan, S, Rezelj, V, Hoppe, N, Boone, M, Billesboelle, C.B, Puchades, C, Azumaya, C.M, Kratochvil, H.T, Zimanyi, M, Desphande, I, Liang, J, Dickinson, S, Nguyen, H.C, Chio, C.M, Merz, G.E, Thompson, M.C, Diwanji, D, Schaefer, K, Anand, A.A, Dobzinski, N, Zha, B.S, Simoneau, C.R, Leon, K, White, K.M, Chio, U.S, Gupta, M, Jin, M, Li, F, Liu, Y, Zhang, K, Bulkley, D, Sun, M, Smith, A.M, Rizo, A.N, Moss, F, Brilot, A.F, Pourmal, S, Trenker, R, Pospiech, T, Gupta, S, Barsi-Rhyne, B, Belyy, V, Barile-Hill, A.W, Nock, S, Liu, Y, Krogan, N.J, Ralston, C.Y, Swaney, D.L, Garcia-Sastre, A, Ott, M, Vignuzzi, M, Walter, P, Manglik, A, QCRG Structural Biology Consortium | | Deposit date: | 2020-10-27 | | Release date: | 2020-11-11 | | Last modified: | 2021-04-21 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | An ultrapotent synthetic nanobody neutralizes SARS-CoV-2 by stabilizing inactive Spike.
Science, 370, 2020
|
|
7KMS
 
 | | Cryo-EM structure of triple ACE2-bound SARS-CoV-2 trimer spike at pH 7.4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Gorman, J, Kwong, P.D, Shapiro, L. | | Deposit date: | 2020-11-03 | | Release date: | 2020-12-09 | | Last modified: | 2021-12-15 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains.
Cell Host Microbe, 28, 2020
|
|
5VK2
 
 | | Structural basis for antibody-mediated neutralization of Lassa virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hastie, K.M, Zandonatti, M.A, Kleinfelter, L.M, Rowland, M.L, Rowland, M.M, Chandra, K, Branco, L.M, Robinson, J.E, Garry, R.F, Saphire, E.O. | | Deposit date: | 2017-04-20 | | Release date: | 2017-05-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.201 Å) | | Cite: | Structural basis for antibody-mediated neutralization of Lassa virus.
Science, 356, 2017
|
|
8BLP
 
 | | Human Urea Transporter UT-B/UT1 in Complex with Inhibitor UTBinh-14 | | Descriptor: | 10-(4-ethylphenyl)sulfonyl-~{N}-(thiophen-2-ylmethyl)-5-thia-1,8,11,12-tetrazatricyclo[7.3.0.0^{2,6}]dodeca-2(6),3,7,9,11-pentaen-7-amine, CHOLESTEROL HEMISUCCINATE, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Chi, G, Dietz, L, Pike, A.C.W, Maclean, E.M, Mukhopadhyay, S.M.M, Bohstedt, T, Wang, D, Scacioc, A, McKinley, G, Arrowsmith, C.H, Edwards, A, Bountra, C, Fernandez-Cid, A, Burgess-Brown, N.A, Duerr, K.L. | | Deposit date: | 2022-11-10 | | Release date: | 2023-10-04 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural characterization of human urea transporters UT-A and UT-B and their inhibition.
Sci Adv, 9, 2023
|
|
4ZKH
 
 | |
7L0V
 
 | |
7L0W
 
 | |
7KE7
 
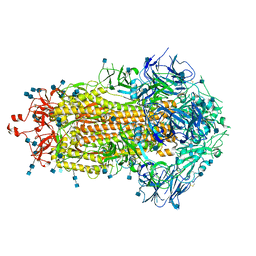 | |
8BWF
 
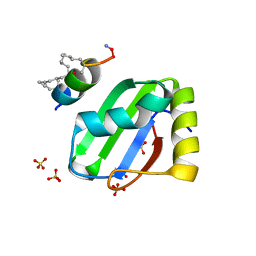 | | PTBP1 RRM1 bound to an allosteric inhibitor | | Descriptor: | AMINO GROUP, GLYCEROL, Ligand, ... | | Authors: | Schmeing, S, Vetter, I, t Hart, P, Gasper, R. | | Deposit date: | 2022-12-06 | | Release date: | 2023-10-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Rationally designed stapled peptides allosterically inhibit PTBP1-RNA-binding.
Chem Sci, 14, 2023
|
|
7KJ4
 
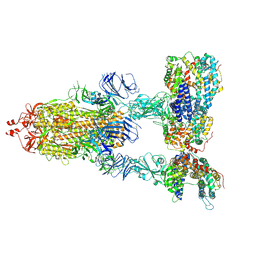 | | SARS-CoV-2 Spike Glycoprotein with three ACE2 Bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Zhang, J, Xiao, T.S, Cai, Y.F, Chen, B. | | Deposit date: | 2020-10-25 | | Release date: | 2020-11-11 | | Last modified: | 2021-02-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | A trimeric human angiotensin-converting enzyme 2 as an anti-SARS-CoV-2 agent.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7KMK
 
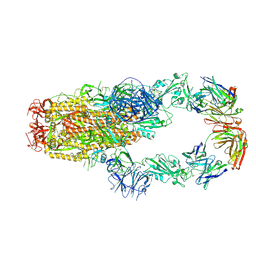 | | cryo-EM structure of SARS-CoV-2 spike in complex with Fab 15033-7, two RBDs bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab 15033-7 heavy chain, ... | | Authors: | Li, Z, Rini, J.M. | | Deposit date: | 2020-11-03 | | Release date: | 2021-02-10 | | Last modified: | 2021-08-25 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Tetravalent SARS-CoV-2 Neutralizing Antibodies Show Enhanced Potency and Resistance to Escape Mutations.
J.Mol.Biol., 433, 2021
|
|
7KMY
 
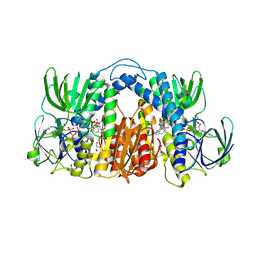 | | Structure of Mtb Lpd bound to 010705 | | Descriptor: | Dihydrolipoyl dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Lima, C.D. | | Deposit date: | 2020-11-03 | | Release date: | 2021-01-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Whole Cell Active Inhibitors of Mycobacterial Lipoamide Dehydrogenase Afford Selectivity over the Human Enzyme through Tight Binding Interactions.
Acs Infect Dis., 7, 2021
|
|
8BAP
 
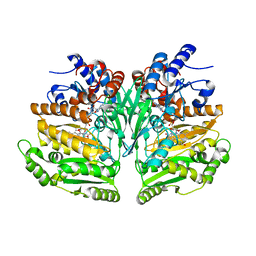 | | Eugenol Oxidase (EUGO) from Rhodococcus jostii RHA1, eightfold mutant active on propanol syringol | | Descriptor: | 4-[(1E)-3-hydroxyprop-1-en-1-yl]-2,6-dimethoxyphenol, CALCIUM ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Alvigini, L, Mattevi, A. | | Deposit date: | 2022-10-11 | | Release date: | 2023-10-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | One-Pot Biocatalytic Synthesis of rac -Syringaresinol from a Lignin-Derived Phenol.
Acs Catalysis, 13, 2023
|
|
7KS9
 
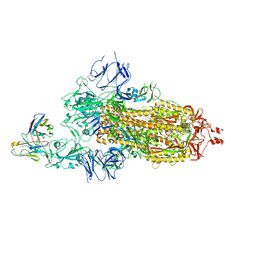 | |
4ZHH
 
 | | Siderocalin-mediated recognition and cellular uptake of actinides | | Descriptor: | CHLORIDE ION, GLYCEROL, N,N'-butane-1,4-diylbis[1-hydroxy-N-(3-{[(1-hydroxy-6-oxo-1,6-dihydropyridin-2-yl)carbonyl]amino}propyl)-6-oxo-1,6-dihydropyridine-2-carboxamide], ... | | Authors: | Allred, B.E, Rupert, P.B, Gauny, S.S, An, D.D, Ralston, C.Y, Sturzbecher-Hoehne, M, Strong, R.K, Abergel, R.J. | | Deposit date: | 2015-04-24 | | Release date: | 2015-08-05 | | Last modified: | 2015-09-02 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Siderocalin-mediated recognition, sensitization, and cellular uptake of actinides.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
7KML
 
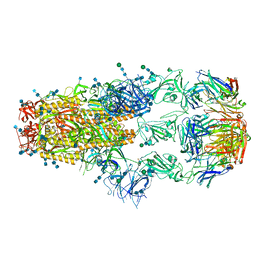 | | cryo-EM structure of SARS-CoV-2 spike in complex with Fab 15033-7, three RBDs bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab 15033-7 heavy chain, ... | | Authors: | Li, Z, Rini, J.M. | | Deposit date: | 2020-11-03 | | Release date: | 2021-02-10 | | Last modified: | 2021-08-25 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Tetravalent SARS-CoV-2 Neutralizing Antibodies Show Enhanced Potency and Resistance to Escape Mutations.
J.Mol.Biol., 433, 2021
|
|
7L2C
 
 | | Crystallographic structure of neutralizing antibody 2-51 in complex with SARS-CoV-2 spike N-terminal domain (NTD) | | Descriptor: | 2-51 heavy chain, 2-51 light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Cerutti, G, Reddem, E.R, Shapiro, L. | | Deposit date: | 2020-12-16 | | Release date: | 2021-02-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | Potent SARS-CoV-2 neutralizing antibodies directed against spike N-terminal domain target a single supersite.
Cell Host Microbe, 29, 2021
|
|
7L6I
 
 | | The empty AAV13 capsid | | Descriptor: | Capsid protein | | Authors: | Mietzsch, M, Agbandje-McKenna, M. | | Deposit date: | 2020-12-23 | | Release date: | 2021-02-03 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Completion of the AAV Structural Atlas: Serotype Capsid Structures Reveals Clade-Specific Features.
Viruses, 13, 2021
|
|
8BCW
 
 | | Photosystem I assembly intermediate of Avena sativa | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Naschberger, A, Amunts, A, Nelson, N. | | Deposit date: | 2022-10-17 | | Release date: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (2.11 Å) | | Cite: | Photosystem I assembly intermediate of Avena sativa
To Be Published
|
|
4ZP3
 
 | | AKAP18:PKA-RIIalpha structure reveals crucial anchor points for recognition of regulatory subunits of PKA | | Descriptor: | A-kinase anchor protein 7 isoforms alpha and beta, CADMIUM ION, cAMP-dependent protein kinase type II-alpha regulatory subunit | | Authors: | Goetz, F, Roske, Y, Faelber, K, Zuehlke, K, Autenrieth, K, Kreuchwig, A, Krause, G, Herberg, F.W, Daumke, O, Heinemann, U, Klussmann, E. | | Deposit date: | 2015-05-07 | | Release date: | 2016-05-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | AKAP18:PKA-RII alpha structure reveals crucial anchor points for recognition of regulatory subunits of PKA.
Biochem.J., 473, 2016
|
|
5W1Y
 
 | | SETD8 in complex with a covalent inhibitor | | Descriptor: | 2-(4-methylpiperazin-1-yl)-3-(phenylsulfanyl)naphthalene-1,4-dione, N-lysine methyltransferase KMT5A, UNKNOWN ATOM OR ION | | Authors: | Tempel, W, Yu, W, Li, Y, Blum, G, Luo, M, Pittella-Silva, F, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-06-05 | | Release date: | 2017-06-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | SETD8 in complex with a covalent inhibitor
to be published
|
|
