6H5X
 
 | | Crystal structure of human Angiotensin-1 converting enzyme N-domain in complex with Omapatrilat. | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Cozier, G.E, Acharya, K.R. | | Deposit date: | 2018-07-25 | | Release date: | 2018-11-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular Basis for Multiple Omapatrilat Binding Sites within the ACE C-Domain: Implications for Drug Design.
J. Med. Chem., 61, 2018
|
|
7NPL
 
 | | ALPHA-1 ANTITRYPSIN (C232S) COMPLEXED WITH cmpd 11 | | Descriptor: | Alpha-1-antitrypsin, GLYCEROL, N-((1S,2R)-1-(3-chloro-2-methylphenyl)-1-hydroxypentan-2-yl)-2-oxoindoline-4-carboxamide | | Authors: | Chung, C. | | Deposit date: | 2021-02-27 | | Release date: | 2021-04-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | The development of highly potent and selective small molecule correctors of Z alpha 1 -antitrypsin misfolding.
Bioorg.Med.Chem.Lett., 41, 2021
|
|
7NPK
 
 | | ALPHA-1 ANTITRYPSIN C232S COMPLEXED WITH CMPD3 | | Descriptor: | Alpha-1-antitrypsin, GLYCEROL, N-((1S,2R)-1-hydroxy-1-(o-tolyl)pentan-2-yl)-2-oxo-2,3-dihydrobenzo[d]oxazole-5-carboxamide | | Authors: | Chung, C. | | Deposit date: | 2021-02-27 | | Release date: | 2021-04-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | The development of highly potent and selective small molecule correctors of Z alpha 1 -antitrypsin misfolding.
Bioorg.Med.Chem.Lett., 41, 2021
|
|
6EKY
 
 | |
6FL8
 
 | | Inositol 1,3,4,5,6-pentakisphosphate 2-kinase from A. thaliana in complex with purpurogallin and ADP | | Descriptor: | 1,2-ETHANEDIOL, 2,3,4,6-tetrahydroxy-5H-benzo[7]annulen-5-one, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Whitfield, H.L, Brearley, C.A, Hemmings, A.M. | | Deposit date: | 2018-01-25 | | Release date: | 2018-09-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Fluorescent Probe Identifies Active Site Ligands of Inositol Pentakisphosphate 2-Kinase.
J. Med. Chem., 61, 2018
|
|
4Z3D
 
 | |
4DB3
 
 | | 1.95 Angstrom Resolution Crystal Structure of N-acetyl-D-glucosamine kinase from Vibrio vulnificus. | | Descriptor: | CHLORIDE ION, GLYCEROL, N-acetyl-D-glucosamine kinase, ... | | Authors: | Minasov, G, Wawrzak, Z, Onopriyenko, O, Skarina, T, Papazisi, L, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-01-13 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | 1.95 Angstrom Resolution Crystal Structure of N-acetyl-D-glucosamine kinase from Vibrio vulnificus.
TO BE PUBLISHED
|
|
5Z49
 
 | | Crystal structure of the effector-binding domain of Synechococcus elongatus CmpR in complex with ribulose-1,5-bisphosphate | | Descriptor: | HTH-type transcriptional activator CmpR, RIBULOSE-1,5-DIPHOSPHATE | | Authors: | Jiang, Y.L, Mahounga, D.M, Sun, H. | | Deposit date: | 2018-01-10 | | Release date: | 2018-10-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.148 Å) | | Cite: | Crystal structure of the effector-binding domain of Synechococcus elongatus CmpR in complex with ribulose 1,5-bisphosphate.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
9DBM
 
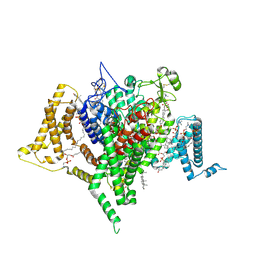 | | Full-length apo human voltage-gated sodium channel 1.8 (NaV1.8), class II | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CHOLESTEROL, ... | | Authors: | Neumann, B, McCarthy, S, Gonen, S. | | Deposit date: | 2024-08-23 | | Release date: | 2025-02-19 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Structural basis of inhibition of human Na V 1.8 by the tarantula venom peptide Protoxin-I.
Nat Commun, 16, 2025
|
|
9DBN
 
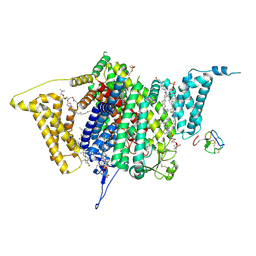 | | Tarantula venom peptide Protoxin-I bound to full-length human voltage-gated sodium channel 1.8 (NaV1.8) | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Beta/omega-theraphotoxin-Tp1a, ... | | Authors: | Neumann, B, McCarthy, S, Gonen, S. | | Deposit date: | 2024-08-23 | | Release date: | 2025-02-19 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Structural basis of inhibition of human Na V 1.8 by the tarantula venom peptide Protoxin-I.
Nat Commun, 16, 2025
|
|
3HJV
 
 | | 1.7 Angstrom resolution crystal structure of an acyl carrier protein S-malonyltransferase from Vibrio cholerae O1 biovar eltor str. N16961 | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, CHLORIDE ION, Malonyl Coa-acyl carrier protein transacylase, ... | | Authors: | Halavaty, A.S, Wawrzak, Z, Anderson, S, Skarina, T, Onopriyenko, O, Kwon, K, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-05-22 | | Release date: | 2009-06-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 1.7 Angstrom resolution crystal structure of an acyl carrier protein S-malonyltransferase from Vibrio cholerae O1 biovar eltor str. N16961
To be Published
|
|
5H9X
 
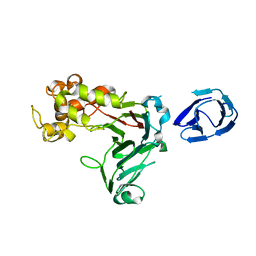 | | Crystal structure of GH family 64 laminaripentaose-producing beta-1,3-glucanase from Paenibacillus barengoltzii | | Descriptor: | beta-1,3-glucanase | | Authors: | Zhen, Q, Yan, Q, Yang, S, Jiang, Z, You, X. | | Deposit date: | 2015-12-29 | | Release date: | 2017-02-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | The recognition mechanism of triple-helical beta-1,3-glucan by a beta-1,3-glucanase
Chem. Commun. (Camb.), 53, 2017
|
|
6D0V
 
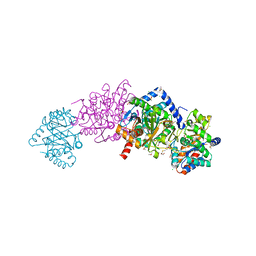 | | Tryptophan synthase Q114A mutant in complex with inhibitor N-(4'-trifluoromethoxybenzenesulfonyl)-2-amino-1-ethylphosphate (F9F) at the alpha-site, aminoacrylate at the beta site, and cesium ion at the metal coordination site | | Descriptor: | 1,2-ETHANEDIOL, 2-({[4-(TRIFLUOROMETHOXY)PHENYL]SULFONYL}AMINO)ETHYL DIHYDROGEN PHOSPHATE, 2-{[(E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}prop-2-enoic acid, ... | | Authors: | Hilario, E, Dunn, M.F, Mueller, L.J, Fan, L. | | Deposit date: | 2018-04-11 | | Release date: | 2019-04-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Tryptophan synthase Q114A mutant in complex with inhibitor N-(4'-trifluoromethoxybenzenesulfonyl)-2-amino-1-ethylphosphate (F9F) at the alpha-site, aminoacrylate at the beta site, and cesium ion at the metal coordination site.
To be Published
|
|
3NUY
 
 | |
5KKB
 
 | | Structure of mouse Golgi alpha-1,2-mannosidase IA and Man9GlcNAc2-PA complex | | Descriptor: | 1,4-BUTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-PYRIDINIUM-1-YLPROPANE-1-SULFONATE, ... | | Authors: | Xiang, Y, Moremen, K.W. | | Deposit date: | 2016-06-21 | | Release date: | 2016-12-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.774 Å) | | Cite: | Substrate recognition and catalysis by GH47 alpha-mannosidases involved in Asn-linked glycan maturation in the mammalian secretory pathway.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
6D4R
 
 | | M. thermoresistible GuaB2 delta-CBS in complex with inhibitor Compound 18 (VCC399134) | | Descriptor: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase,Inosine-5'-monophosphate dehydrogenase, hydroxy(3-{4-[(isoquinolin-5-yl)sulfonyl]piperazine-1-carbonyl}phenyl)oxoammonium | | Authors: | Ascher, D.B, Pacitto, A, Blundell, T.L. | | Deposit date: | 2018-04-18 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Synthesis and Structure-Activity relationship of 1-(5-isoquinolinesulfonyl)piperazine analogues as inhibitors of Mycobacterium tuberculosis IMPDH.
Eur.J.Med.Chem., 174, 2019
|
|
5Z90
 
 | | BRD4 Bromodomain 1 with an inhibitor | | Descriptor: | 1-ethyl-6-pyrrolidin-1-ylsulfonyl-benzo[cd]indol-2-one, Bromodomain-containing protein 4 | | Authors: | Xiao, S, Chen, S, Chen, H. | | Deposit date: | 2018-02-01 | | Release date: | 2019-02-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | BRD4 Bromodomain 1 with an inhibitor
To Be Published
|
|
6DB5
 
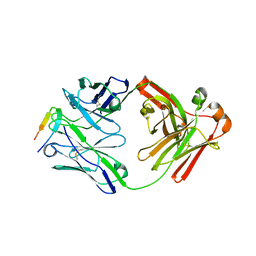 | |
6D4T
 
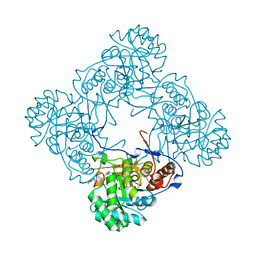 | | M. thermoresistible GuaB2 delta-CBS in complex with inhibitor Compound 45 (VCC117054) | | Descriptor: | (2S)-N-(2H-1,3-benzodioxol-5-yl)-4-[(isoquinolin-5-yl)sulfonyl]-2-methylpiperazine-1-carboxamide, INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase,Inosine-5'-monophosphate dehydrogenase | | Authors: | Ascher, D.B, Pacitto, A, Blundell, T.L. | | Deposit date: | 2018-04-18 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Synthesis and Structure-Activity relationship of 1-(5-isoquinolinesulfonyl)piperazine analogues as inhibitors of Mycobacterium tuberculosis IMPDH.
Eur.J.Med.Chem., 174, 2019
|
|
2A5R
 
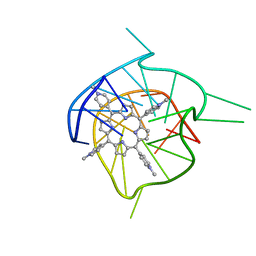 | | Complex of tetra-(4-n-methylpyridyl) porphin with monomeric parallel-stranded DNA tetraplex, snap-back 3+1 3' G-tetrad, single-residue chain reversal loops, GAG triad in the context of GAAG diagonal loop, C-MYC promoter, NMR, 6 struct. | | Descriptor: | (1Z,4Z,9Z,15Z)-5,10,15,20-tetrakis(1-methylpyridin-1-ium-4-yl)-21,23-dihydroporphyrin, 5'-D(*TP*GP*AP*GP*GP*GP*TP*GP*GP*IP*GP*AP*GP*GP*GP*TP*GP*GP*GP*GP*AP*AP*GP*G)-3' | | Authors: | Phan, A.T, Kuryavyi, V.V, Gaw, H.Y, Patel, D.J. | | Deposit date: | 2005-06-30 | | Release date: | 2005-07-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Small-molecule interaction with a five-guanine-tract G-quadruplex structure from the human MYC promoter.
Nat.Chem.Biol., 1, 2005
|
|
6D4S
 
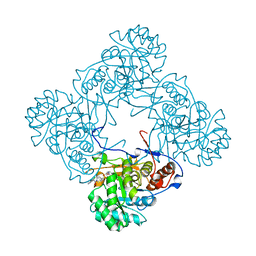 | | M. thermoresistible GuaB2 delta-CBS in complex with inhibitor Compound 37 (VCC670597) | | Descriptor: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase,Inosine-5'-monophosphate dehydrogenase, N-(2,3-dichlorophenyl)-4-[(isoquinolin-5-yl)sulfonyl]piperazine-1-carboxamide | | Authors: | Ascher, D.B, Pacitto, A, Blundell, T.L. | | Deposit date: | 2018-04-18 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Synthesis and Structure-Activity relationship of 1-(5-isoquinolinesulfonyl)piperazine analogues as inhibitors of Mycobacterium tuberculosis IMPDH.
Eur.J.Med.Chem., 174, 2019
|
|
5Z8G
 
 | | BRD4 Bromodomain 1 with an inhibitor | | Descriptor: | 1-ethyl-6-[(3R)-3-oxidanylpiperidin-1-yl]sulfonyl-benzo[cd]indol-2-one, Bromodomain-containing protein 4, DIMETHYL SULFOXIDE | | Authors: | Xiao, S, Chen, S, Chen, H. | | Deposit date: | 2018-01-31 | | Release date: | 2019-02-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | BRD4 Bromodomain 1 with an inhibitor
To Be Published
|
|
5FDO
 
 | | Mcl-1 complexed with small molecule inhibitor | | Descriptor: | 3-[3-(4-chloranyl-3,5-dimethyl-phenoxy)propyl]-~{N}-(phenylsulfonyl)-1~{H}-indole-2-carboxamide, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Zhao, B. | | Deposit date: | 2015-12-16 | | Release date: | 2016-03-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of 2-Indole-acylsulfonamide Myeloid Cell Leukemia 1 (Mcl-1) Inhibitors Using Fragment-Based Methods.
J.Med.Chem., 59, 2016
|
|
4LH3
 
 | |
4LH1
 
 | |
