8R2I
 
 | | Cryo-EM Structure of native Photosystem II assembly intermediate from Chlamydomonas reinhardtii | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, 2,3-DIMETHYL-5-(3,7,11,15,19,23,27,31,35-NONAMETHYL-2,6,10,14,18,22,26,30,34-HEXATRIACONTANONAENYL-2,5-CYCLOHEXADIENE-1,4-DIONE-2,3-DIMETHYL-5-SOLANESYL-1,4-BENZOQUINONE, ... | | Authors: | Fadeeva, M, Klaiman, D, Kandiah, E, Nelson, N. | | Deposit date: | 2023-11-06 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure of native photosystem II assembly intermediate from Chlamydomonas reinhardtii .
Front Plant Sci, 14, 2023
|
|
8R16
 
 | | Structure of compound 12 bound to SARS-CoV-2 main protease | | Descriptor: | 1,2-ETHANEDIOL, 1-[6,7-bis(chloranyl)-3,4-dihydro-1H-isoquinolin-2-yl]-2-(5-methylpyridin-3-yl)ethanone, 3C-like proteinase, ... | | Authors: | Mac Sweeney, A, Hazemann, J. | | Deposit date: | 2023-11-01 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Identification of SARS-CoV-2 Mpro inhibitors through deep reinforcement learning for de novo drug design and computational chemistry approaches
To Be Published
|
|
8R12
 
 | | Structure of compound 8 bound to SARS-CoV-2 main protease | | Descriptor: | 2-[[4-(5-chloranylpyridin-3-yl)carbonyl-1,4-diazepan-1-yl]methyl]benzenecarbonitrile, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Mac Sweeney, A, Hazemann, J. | | Deposit date: | 2023-11-01 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.587 Å) | | Cite: | Identification of SARS-CoV-2 Mpro inhibitors through deep reinforcement learning for de novo drug design and computational chemistry approaches
To Be Published
|
|
8R14
 
 | | Structure of compound 11 bound to SARS-CoV-2 main protease | | Descriptor: | (5-chloranylpyridin-3-yl)-[4-[(2-chlorophenyl)methyl]-1,4-diazepan-1-yl]methanone, 3C-like proteinase, BROMIDE ION, ... | | Authors: | Mac Sweeney, A, Hazemann, J. | | Deposit date: | 2023-11-01 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.336 Å) | | Cite: | Identification of SARS-CoV-2 Mpro inhibitors through deep reinforcement learning for de novo drug design and computational chemistry approaches
To Be Published
|
|
8R11
 
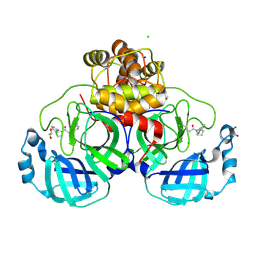 | | Structure of compound 7 bound to SARS-CoV-2 main protease | | Descriptor: | 1,2-ETHANEDIOL, 1-[(2~{S})-2-(3-chlorophenyl)pyrrolidin-1-yl]-2-(5-methylpyridin-3-yl)ethanone, 3C-like proteinase, ... | | Authors: | Mac Sweeney, A, Hazemann, J. | | Deposit date: | 2023-11-01 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Identification of SARS-CoV-2 Mpro inhibitors through deep reinforcement learning for de novo drug design and computational chemistry approaches
To Be Published
|
|
8R1D
 
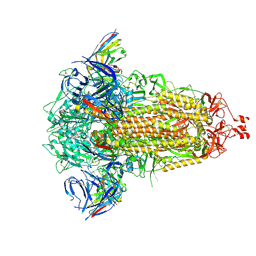 | | SD1-3 Fab in complex with SARS-CoV-2 BA.2.12.1 Spike Glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SD1-3 Fab Heavy Chain, SD1-3 Fab Light Chain, ... | | Authors: | Duyvesteyn, H.M.E, Ren, J, Stuart, D.I. | | Deposit date: | 2023-11-01 | | Release date: | 2024-03-13 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (2.37 Å) | | Cite: | The SARS-CoV-2 neutralizing antibody response to SD1 and its evasion by BA.2.86.
Nat Commun, 15, 2024
|
|
8UU4
 
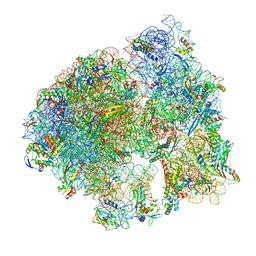 | | Cryo-EM structure of the Listeria innocua 70S ribosome in complex with HPF (structure I-A) | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 5S Ribosomal RNA, ... | | Authors: | Seely, S.M, Basu, R.S, Gagnon, M.G. | | Deposit date: | 2023-10-31 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Mechanistic insights into the alternative ribosome recycling by HflXr.
Nucleic Acids Res., 52, 2024
|
|
8UU8
 
 | | Cryo-EM structure of the Listeria innocua 70S ribosome (head-swiveled) in complex with HflXr and pe/E-tRNA (structure II-C) | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 5S Ribosomal RNA, ... | | Authors: | Seely, S.M, Basu, R.S, Gagnon, M.G. | | Deposit date: | 2023-10-31 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Mechanistic insights into the alternative ribosome recycling by HflXr.
Nucleic Acids Res., 52, 2024
|
|
8UU5
 
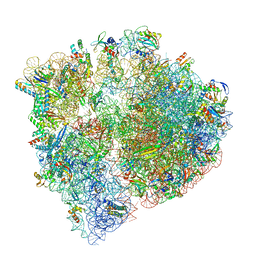 | | Cryo-EM structure of the Listeria innocua 70S ribosome (head-swiveled) in complex with pe/E-tRNA (structure I-B) | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 5S Ribosomal RNA, ... | | Authors: | Seely, S.M, Basu, R.S, Gagnon, M.G. | | Deposit date: | 2023-10-31 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Mechanistic insights into the alternative ribosome recycling by HflXr.
Nucleic Acids Res., 52, 2024
|
|
8UUA
 
 | | Cryo-EM structure of the Listeria innocua 50S ribosomal subunit in complex with HflXr (structure III) | | Descriptor: | 23S Ribosomal RNA, 5S Ribosomal RNA, GTPase HflX, ... | | Authors: | Seely, S.M, Basu, R.S, Gagnon, M.G. | | Deposit date: | 2023-10-31 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Mechanistic insights into the alternative ribosome recycling by HflXr.
Nucleic Acids Res., 52, 2024
|
|
8UU6
 
 | | Cryo-EM structure of the ratcheted Listeria innocua 70S ribosome in complex with p/E-tRNA (structure II-A) | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 5S Ribosomal RNA, ... | | Authors: | Seely, S.M, Basu, R.S, Gagnon, M.G. | | Deposit date: | 2023-10-31 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Mechanistic insights into the alternative ribosome recycling by HflXr.
Nucleic Acids Res., 52, 2024
|
|
8UU7
 
 | | Cryo-EM structure of the Listeria innocua 70S ribosome in complex with HflXr, HPF, and E-site tRNA (structure II-B) | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 5S Ribosomal RNA, ... | | Authors: | Seely, S.M, Basu, R.S, Gagnon, M.G. | | Deposit date: | 2023-10-31 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mechanistic insights into the alternative ribosome recycling by HflXr.
Nucleic Acids Res., 52, 2024
|
|
8UU9
 
 | | Cryo-EM structure of the ratcheted Listeria innocua 70S ribosome (head-swiveled) in complex with HflXr and pe/E-tRNA (structure II-D) | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 5S Ribosomal RNA, ... | | Authors: | Seely, S.M, Basu, R.S, Gagnon, M.G. | | Deposit date: | 2023-10-31 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Mechanistic insights into the alternative ribosome recycling by HflXr.
Nucleic Acids Res., 52, 2024
|
|
8R0B
 
 | | Cryo-EM structure of the cross-exon pre-B+ATP complex | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, 5'ss oligo, NHP2-like protein 1, ... | | Authors: | Zhang, Z, Kumar, V, Dybkov, O, Will, C.L, Zhong, J, Ludwig, S, Urlaub, H, Kastner, B, Stark, H, Luehrmann, R. | | Deposit date: | 2023-10-31 | | Release date: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural insights into the cross-exon to cross-intron spliceosome switch
Nature, 2024
|
|
8R08
 
 | | Cryo-EM structure of the cross-exon pre-B+AMPPNP complex | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, NHP2-like protein 1, N-terminally processed, ... | | Authors: | Zhang, Z, Kumar, V, Dybkov, O, Will, C.L, Zhong, J, Ludwig, S, Urlaub, H, Kastner, B, Stark, H, Luehrmann, R. | | Deposit date: | 2023-10-31 | | Release date: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Structural insights into the cross-exon to cross-intron spliceosome switch
Nature, 2024
|
|
8R0A
 
 | | Cryo-EM structure of the cross-exon pre-B+5'ss complex | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, 5'ss oligo, NHP2-like protein 1, ... | | Authors: | Zhang, Z, Kumar, V, Dybkov, O, Will, C.L, Zhong, J, Ludwig, S, Urlaub, H, Kastner, B, Stark, H, Luehrmann, R. | | Deposit date: | 2023-10-31 | | Release date: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | Structural insights into the cross-exon to cross-intron spliceosome switch
Nature, 2024
|
|
8R09
 
 | | Cryo-EM structure of the cross-exon pre-B+5'ss+ATPgammaS complex | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, 5'ss oligo, NHP2-like protein 1, ... | | Authors: | Zhang, Z, Kumar, V, Dybkov, O, Will, C.L, Zhong, J, Ludwig, S, Urlaub, H, Kastner, B, Stark, H, Luehrmann, R. | | Deposit date: | 2023-10-31 | | Release date: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural insights into the cross-exon to cross-intron spliceosome switch
Nature, 2024
|
|
8UT1
 
 | | Alpha7-nicotinic acetylcholine receptor bound to epibatidine | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Burke, S.M, Hibbs, R.E, Noviello, C.M. | | Deposit date: | 2023-10-30 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Structural mechanisms of alpha 7 nicotinic receptor allosteric modulation and activation.
Cell, 187, 2024
|
|
8UTB
 
 | | Alpha7-nicotinic acetylcholine receptor bound to epibatidine and NS-1738 | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Burke, S.M, Noviello, C.M, Hibbs, R.E. | | Deposit date: | 2023-10-30 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Structural mechanisms of alpha 7 nicotinic receptor allosteric modulation and activation.
Cell, 187, 2024
|
|
8R04
 
 | | Structure of Staphylococcus aureus ClpP Bound to the Covalent Active Site Inhibitor Cystargolide A | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, Cystargolide A (bound) | | Authors: | Illigmann, A, Vielberg, M.-T, Lakemeyer, M, Wolf, F, Staudt, N, Dema, T, Stange, P, Liebhart, E, Kuttenlochner, W, Kulik, A, Malik, I, Grond, S, Sieber, S.A, Groll, M, Kaysser, L, Broetz-Oesterhelt, H. | | Deposit date: | 2023-10-30 | | Release date: | 2023-12-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of Staphylococcus aureus ClpP Bound to the Covalent Active-Site Inhibitor Cystargolide A.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8R05
 
 | | Photorhabdus lamondii ClpP in complex with the natural product beta-lactone inhibitor Cystargolide A at 2.5 A resolution | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, Cystargolide A (bound) | | Authors: | Illigmann, A, Vielberg, M.-T, Lakemeyer, M, Wolf, F, Staudt, N, Dema, T, Stange, P, Liebhart, E, Kuttenlochner, W, Kulik, A, Malik, I, Grond, S, Sieber, S.A, Groll, M, Kaysser, L, Broetz-Oesterhelt, H. | | Deposit date: | 2023-10-30 | | Release date: | 2023-12-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of Staphylococcus aureus ClpP Bound to the Covalent Active-Site Inhibitor Cystargolide A.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8R03
 
 | | Staphylococcus aureus ClpP in complex with the natural product beta-lactone inhibitor Cystargolide A at 2.0 A resolution | | Descriptor: | 1,2-ETHANEDIOL, ATP-dependent Clp protease proteolytic subunit, Cystargolide A (bound) | | Authors: | Illigmann, A, Vielberg, M.-T, Lakemeyer, M, Wolf, F, Staudt, N, Dema, T, Stange, P, Liebhart, E, Kuttenlochner, W, Kulik, A, Malik, I, Grond, S, Sieber, S.A, Groll, M, Kaysser, L, Broetz-Oesterhelt, H. | | Deposit date: | 2023-10-30 | | Release date: | 2023-12-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Staphylococcus aureus ClpP Bound to the Covalent Active-Site Inhibitor Cystargolide A.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8QZS
 
 | | Cryo-EM structure of the cross-exon B-like complex | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, Microfibrillar-associated protein 1, NHP2-like protein 1, ... | | Authors: | Zhang, Z, Kumar, V, Dybkov, O, Will, C.L, Zhong, J, Ludwig, S, Urlaub, H, Kastner, B, Stark, H, Luehrmann, R. | | Deposit date: | 2023-10-29 | | Release date: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural insights into the cross-exon to cross-intron spliceosome switch
Nature, 2024
|
|
8WXB
 
 | | Cryo-EM structure of the alpha-carboxysome shell vertex from Prochlorococcus MED4 | | Descriptor: | Carboxysome assembly protein CsoS2, Carboxysome shell vertex protein CsoS4A, Major carboxysome shell protein CsoS1 | | Authors: | Jiang, Y.L, Zhou, R.Q, Zhou, C.Z, Zeng, Q.L. | | Deposit date: | 2023-10-28 | | Release date: | 2024-01-31 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structure and assembly of the alpha-carboxysome in the marine cyanobacterium Prochlorococcus.
Nat.Plants, 10, 2024
|
|
8URX
 
 | | Escherichia coli transcription-translation coupled complex class B (TTC-B) containing RfaH bound to ops signal, mRNA with a 30 nt long spacer, and fMet-tRNAs in E-site and P-site of the ribosome | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Molodtsov, V, Wang, C, Ebright, R.H. | | Deposit date: | 2023-10-27 | | Release date: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Escherichia coli transcription-translation coupled complex class A (TTC-A) containing RfaH bound to ops signal, mRNA with a 21 nt long spacer, and fMet-tRNAs in E-site and P-site of the ribosome
Nat.Struct.Mol.Biol., 2024
|
|
