1IMA
 
 | |
3C33
 
 | |
4LKR
 
 | | Crystal structure of deoD-3 gene product from Shewanella oneidensis MR-1, NYSGRC target 029437 | | Descriptor: | Purine nucleoside phosphorylase DeoD-type | | Authors: | Malashkevich, V.N, Bonanno, J.B, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-07-08 | | Release date: | 2013-07-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of deoD-3 gene product from Shewanella oneidensis MR-1, NYSGRC target 029437
To be Published
|
|
2QT1
 
 | | Human nicotinamide riboside kinase 1 in complex with nicotinamide riboside | | Descriptor: | Nicotinamide riboside, Nicotinamide riboside kinase 1, PHOSPHATE ION, ... | | Authors: | Rabeh, W.M, Tempel, W, Nedyalkova, L, Landry, R, Arrowsmith, C.H, Edwards, A.M, Sundstrom, M, Weigelt, J, Bochkarev, A, Brenner, C, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-07-31 | | Release date: | 2007-08-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Nicotinamide Riboside Kinase Structures Reveal New Pathways to NAD(+).
Plos Biol., 5, 2007
|
|
2XUK
 
 | | CRYSTAL STRUCTURE OF MACHE-Y337A-TZ2PA6 SYN COMPLEX (10 MTH) | | Descriptor: | 3,8-DIAMINO-6-PHENYL-5-[6-[1-[2-[(1,2,3,4-TETRAHYDRO-9-ACRIDINYL)AMINO]ETHYL]-1H-1,2,3-TRIAZOL-5-YL]HEXYL]-PHENANTHRIDINIUM, ACETYLCHOLINESTERASE | | Authors: | Bourne, Y, Radic, Z, Taylor, P, Marchot, P. | | Deposit date: | 2010-10-19 | | Release date: | 2010-12-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Conformational Remodeling of Femtomolar Inhibitor-Acetylcholinesterase Complexes in the Crystalline State
J.Am.Chem.Soc., 132, 2010
|
|
3FQL
 
 | | Hepatitis C virus polymerase NS5B (CON1 1-570) with HCV-796 inhibitor | | Descriptor: | 5-cyclopropyl-2-(4-fluorophenyl)-6-[(2-hydroxyethyl)(methylsulfonyl)amino]-N-methyl-1-benzofuran-3-carboxamide, GLYCEROL, RNA-directed RNA polymerase | | Authors: | Harris, S.F, Wong, A. | | Deposit date: | 2009-01-07 | | Release date: | 2009-02-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Slow binding inhibition and mechanism of resistance of non-nucleoside polymerase inhibitors of hepatitis C virus.
J.Biol.Chem., 284, 2009
|
|
4OU1
 
 | | Crystal structure of a computationally designed retro-aldolase covalently bound to folding probe 1 [(6-methoxynaphthalen-2-yl)(oxiran-2-yl)methanol] | | Descriptor: | (1S,2S)-1-(6-methoxynaphthalen-2-yl)propane-1,2-diol, BENZOIC ACID, PHOSPHATE ION, ... | | Authors: | Bhabha, G, Zhang, X, Ekiert, D.C. | | Deposit date: | 2014-02-14 | | Release date: | 2014-03-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Small molecule probes to quantify the functional fraction of a specific protein in a cell with minimal folding equilibrium shifts.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4HD9
 
 | | Crystal structure of native human MAdCAM-1 D1D2 domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Mucosal addressin cell adhesion molecule 1 | | Authors: | Springer, T, Yu, Y, Zhu, J. | | Deposit date: | 2012-10-02 | | Release date: | 2013-01-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A Different Fold with an Integrin-Binding Loop Specialized for Flexibility in Mucosal Addressin Cell Adhesion Molecule-1
TO BE PUBLISHED
|
|
5RF2
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with Z1741969146 | | Descriptor: | 1-azanylpropylideneazanium, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-03-15 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
1NMK
 
 | | The Sanglifehrin-Cyclophilin Interaction: Degradation Work, Synthetic Macrocyclic Analogues, X-ray Crystal Structure and Binding Data | | Descriptor: | (13E,15E)-(3S,6S,9R,10R,11S,12S,18S,21S)-10,12-DIHYDROXY-3-(3-HYDROXYBEN-ZYL)-18-((E)-3-HYDROXY-1-METHYLPROPENYL)-6-ISOPROPYL-11-METHYL-9-(3-OXO-BUTYL)-19-OXA-1,4,7,25-TETRAAZA-BICYCLO[19.3.1]PENTACOSA-13,15-DIENE-2,5,8,20-TETRAONE, Peptidyl-prolyl cis-trans isomerase A | | Authors: | Kallen, J, Sedrani, R, Wagner, J. | | Deposit date: | 2003-01-10 | | Release date: | 2003-04-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Sanglifehrin-Cyclophilin Interaction: Degradation Work, Synthetic Macrocyclic Analogues, X-ray Crystal Structure and Binding Data
J.Am.Chem.Soc., 125, 2003
|
|
4OWM
 
 | | Anthranilate phosphoribosyl transferase from Mycobacterium tuberculosis in complex with 3-fluoroanthranilate, PRPP and Magnesium | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, 2-azanyl-3-fluoranyl-benzoic acid, Anthranilate phosphoribosyltransferase, ... | | Authors: | Castell, A, Cookson, T.V.M, Short, F.L, Lott, J.S. | | Deposit date: | 2014-02-02 | | Release date: | 2014-04-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Alternative substrates reveal catalytic cycle and key binding events in the reaction catalysed by anthranilate phosphoribosyltransferase from Mycobacterium tuberculosis.
Biochem.J., 461, 2014
|
|
4OF8
 
 | | Crystal Structure of Rst D1-D2 | | Descriptor: | GLYCEROL, Irregular chiasm C-roughest protein, SODIUM ION | | Authors: | Ozkan, E, Garcia, K.C. | | Deposit date: | 2014-01-14 | | Release date: | 2014-02-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Extracellular Architecture of the SYG-1/SYG-2 Adhesion Complex Instructs Synaptogenesis.
Cell(Cambridge,Mass.), 156, 2014
|
|
3C36
 
 | |
2PSG
 
 | |
2V0H
 
 | | Characterization of Substrate Binding and Catalysis of the Potential Antibacterial Target N-acetylglucosamine-1-phosphate Uridyltransferase (GlmU) | | Descriptor: | BIFUNCTIONAL PROTEIN GLMU, COBALT (II) ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Mochalkin, I, Lightle, S, Ohren, J.F, Chirgadze, N.Y. | | Deposit date: | 2007-05-14 | | Release date: | 2008-01-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Characterization of Substrate Binding and Catalysis in the Potential Antibacterial Target N-Acetylglucosamine-1-Phosphate Uridyltransferase (Glmu).
Protein Sci., 16, 2007
|
|
1RL8
 
 | | Crystal structure of the complex of resistant strain of hiv-1 protease(v82a mutant) with ritonavir | | Descriptor: | RITONAVIR, protease RETROPEPSIN | | Authors: | Rezacova, P, Brynda, J, Sedlacek, J, Konvalinka, J, Fabry, M, Horejsi, M. | | Deposit date: | 2003-11-25 | | Release date: | 2005-04-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the complex of resistant strain of hiv-1 protease(v82a mutant) with ritonavir
To be Published, 2005
|
|
7L25
 
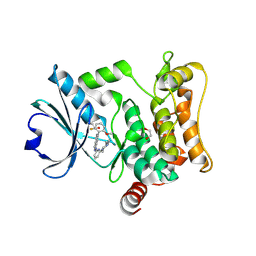 | | HPK1 IN COMPLEX WITH COMPOUND 18 | | Descriptor: | 1,2-ETHANEDIOL, 6-(2-fluoro-6-methoxyphenyl)-1-[6-(4-methylpiperazin-1-yl)pyridin-2-yl]-1H-pyrazolo[4,3-c]pyridine, Mitogen-activated protein kinase kinase kinase kinase 1 | | Authors: | Lesburg, C.A. | | Deposit date: | 2020-12-16 | | Release date: | 2021-03-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Identification of Potent Reverse Indazole Inhibitors for HPK1.
Acs Med.Chem.Lett., 12, 2021
|
|
3FS2
 
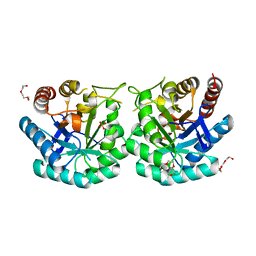 | |
4AYO
 
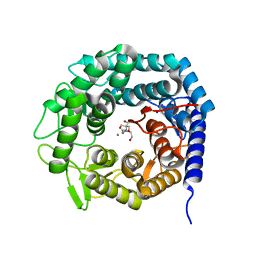 | | Structure of The GH47 processing alpha-1,2-mannosidase from Caulobacter strain K31 | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, MANNOSYL-OLIGOSACCHARIDE 1,2-ALPHA-MANNOSIDASE, ... | | Authors: | Thompson, A.J, Dabin, J, Iglesias-Fernandez, J, Iglesias-Fernandez, A, Dinev, Z, Williams, S.J, Siriwardena, A, Moreland, C, Hu, T.C, Smith, D.K, Gilbert, H.J, Rovira, C, Davies, G.J. | | Deposit date: | 2012-06-21 | | Release date: | 2013-01-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | The Reaction Coordinate of a Bacterial Gh47 Alpha-Mannosidase: A Combined Quantum Mechanical and Structural Approach.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
1JLS
 
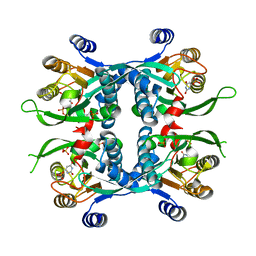 | | STRUCTURE OF THE URACIL PHOSPHORIBOSYLTRANSFERASE URACIL/CPR 2 MUTANT C128V | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Schumacher, M.A, Bashor, C.J, Otsu, K, Zu, S, Parry, R, Ullman, B, Brennan, R.G. | | Deposit date: | 2001-07-16 | | Release date: | 2002-01-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structural mechanism of GTP stabilized oligomerization and catalytic activation of the Toxoplasma gondii uracil phosphoribosyltransferase.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1JOC
 
 | | EEA1 homodimer of C-terminal FYVE domain bound to inositol 1,3-diphosphate | | Descriptor: | Early Endosomal Autoantigen 1, PHOSPHORIC ACID MONO-(2,3,4,6-TETRAHYDROXY-5-PHOSPHONOOXY-CYCLOHEXYL) ESTER, ZINC ION | | Authors: | Dumas, J.J, Merithew, E, Rajamani, D, Hayes, S, Lawe, D, Corvera, S, Lambright, D.G. | | Deposit date: | 2001-07-27 | | Release date: | 2001-12-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Multivalent endosome targeting by homodimeric EEA1.
Mol.Cell, 8, 2001
|
|
3O6G
 
 | | Ligand-binding domain of GluA2 (flip) ionotropic glutamate receptor in complex with an allosteric modulator | | Descriptor: | 1,2-ETHANEDIOL, GLUTAMIC ACID, Glutamate receptor 2, ... | | Authors: | Maclean, J.K.F, Campbell, R.A, Cumming, I.A, Gillen, K.J, Gillespie, J, Jamieson, C, Kazemier, B, Kiczun, M, Lamont, Y, Lyons, A.J, Martin, F, Moir, E.M, Morrow, J.A, Pantling, J, Rankovic, Z, Smith, L. | | Deposit date: | 2010-07-29 | | Release date: | 2010-09-15 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A novel series of positive modulators of the AMPA receptor: structure-based lead optimization.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3SC1
 
 | | Novel Isoquinolone PDK1 Inhibitors Discovered through Fragment-Based Lead Discovery | | Descriptor: | 3-phosphoinositide-dependent protein kinase 1, 6-[2-(hydroxymethyl)phenyl]isoquinolin-1(2H)-one, GLYCEROL, ... | | Authors: | Greasley, S.E, Ferre, R.-A, Krauss, M, Cronin, C. | | Deposit date: | 2011-06-06 | | Release date: | 2011-10-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Novel isoquinolone PDK1 inhibitors discovered through fragment-based lead discovery.
J Comput Aided Mol Des, 25, 2011
|
|
5IFP
 
 | | STRUCTURE OF BETA-GALACTOSIDASE FROM ASPERGILLUS NIGER | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Rico-Diaz, A, Ramirez-Escudero, M, Vizoso Vazquez, A, Cerdan, M.E, Becerra, M, Sanz-Aparicio, J. | | Deposit date: | 2016-02-26 | | Release date: | 2017-04-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Structural features of Aspergillus niger beta-galactosidase define its activity against glycoside linkages.
FEBS J., 284, 2017
|
|
3SL4
 
 | | Crystal structure of the catalytic domain of PDE4D2 with compound 10D | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Feil, S.F. | | Deposit date: | 2011-06-24 | | Release date: | 2011-10-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Thiophene inhibitors of PDE4: Crystal structures show a second binding mode at the catalytic domain of PDE4D2.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
