8JKN
 
 | |
8JKO
 
 | |
7K3R
 
 | | Cryo-EM structure of AIM2-PYD filament | | Descriptor: | Interferon-inducible protein AIM2 | | Authors: | Zheng, W, Matyszewski, M, Sohn, J, Egelman, E.H. | | Deposit date: | 2020-09-12 | | Release date: | 2021-05-26 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Distinct axial and lateral interactions within homologous filaments dictate the signaling specificity and order of the AIM2-ASC inflammasome.
Nat Commun, 12, 2021
|
|
1AHM
 
 | | DER F 2, THE MAJOR MITE ALLERGEN FROM DERMATOPHAGOIDES FARINAE, NMR, 10 STRUCTURES | | Descriptor: | DER F 2 | | Authors: | Ichikawa, S, Hatanaka, H, Yuuki, T, Iwamoto, N, Ogura, K, Okumura, Y, Inagaki, F. | | Deposit date: | 1997-04-07 | | Release date: | 1998-04-08 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Der f 2, the major mite allergen for atopic diseases.
J.Biol.Chem., 273, 1998
|
|
1AHK
 
 | | DER F 2, THE MAJOR MITE ALLERGEN FROM DERMATOPHAGOIDES FARINAE, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | DER F 2 | | Authors: | Ichikawa, S, Hatanaka, H, Yuuki, T, Iwamoto, N, Ogura, K, Okumura, Y, Inagaki, F. | | Deposit date: | 1997-04-07 | | Release date: | 1998-04-08 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Der f 2, the major mite allergen for atopic diseases.
J.Biol.Chem., 273, 1998
|
|
7X45
 
 | | Grass carp interferon gamma related | | Descriptor: | Interferon gamma | | Authors: | Wang, J, Zou, J, Zhu, X. | | Deposit date: | 2022-03-02 | | Release date: | 2022-09-14 | | Last modified: | 2023-04-19 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Novel Dimeric Architecture of an IFN-gamma-Related Cytokine Provides Insights into Subfunctionalization of Type II IFNs in Teleost Fish.
J Immunol., 209, 2022
|
|
7DNJ
 
 | | K63-polyUb MDA5CARDs complex | | Descriptor: | Interferon-induced helicase C domain-containing protein 1, Ubiquitin | | Authors: | Song, B, Chen, Y, Luo, D.H, Zheng, J. | | Deposit date: | 2020-12-09 | | Release date: | 2021-10-13 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Ordered assembly of the cytosolic RNA-sensing MDA5-MAVS signaling complex via binding to unanchored K63-linked poly-ubiquitin chains.
Immunity, 54, 2021
|
|
7DNI
 
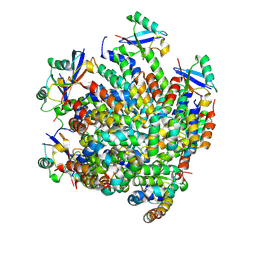 | | MDA5 CARDs-MAVS CARD polyUb complex | | Descriptor: | Interferon-induced helicase C domain-containing protein 1, Mitochondrial antiviral-signaling protein, Ubiquitin | | Authors: | Song, B, Chen, Y, Luo, D.H, Zheng, J. | | Deposit date: | 2020-12-09 | | Release date: | 2021-10-13 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Ordered assembly of the cytosolic RNA-sensing MDA5-MAVS signaling complex via binding to unanchored K63-linked poly-ubiquitin chains.
Immunity, 54, 2021
|
|
7WKH
 
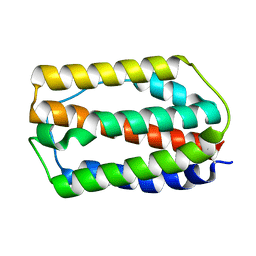 | | the grass carp IFNa | | Descriptor: | Interferon | | Authors: | Zou, J, WANG, J, Wang, Z. | | Deposit date: | 2022-01-09 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural and Functional Analyses of Type I IFNa Shed Light Into Its Interaction With Multiple Receptors in Fish.
Front Immunol, 13, 2022
|
|
3LJB
 
 | |
3SZR
 
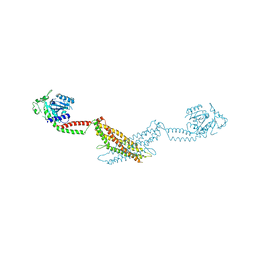 | | Crystal structure of modified nucleotide-free human MxA | | Descriptor: | Interferon-induced GTP-binding protein Mx1 | | Authors: | Gao, S, Daumke, O. | | Deposit date: | 2011-07-19 | | Release date: | 2011-10-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of myxovirus resistance protein a reveals intra- and intermolecular domain interactions required for the antiviral function.
Immunity, 35, 2011
|
|
6C6K
 
 | | Structural basis for preferential recognition of cap 0 RNA by a human IFIT1-IFIT3 protein complex | | Descriptor: | Interferon-induced protein with tetratricopeptide repeats 1, Interferon-induced protein with tetratricopeptide repeats 3, MAGNESIUM ION, ... | | Authors: | Amarasinghe, G.K, Leung, D.W, Johnson, B, Xu, W. | | Deposit date: | 2018-01-18 | | Release date: | 2018-04-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Human IFIT3 Modulates IFIT1 RNA Binding Specificity and Protein Stability.
Immunity, 48, 2018
|
|
3M66
 
 | | Crystal structure of human Mitochondrial Transcription Termination Factor 3 | | Descriptor: | mTERF domain-containing protein 1, mitochondrial | | Authors: | Spahr, H, Samuelsson, T, Hallberg, B.M, Gustafsson, C.M. | | Deposit date: | 2010-03-15 | | Release date: | 2010-05-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of mitochondrial transcription termination factor 3 reveals a novel nucleic acid-binding domain.
Biochem.Biophys.Res.Commun., 397, 2010
|
|
3UX9
 
 | | Structural insights into a human anti-IFN antibody exerting therapeutic potential for systemic lupus erythematosus | | Descriptor: | Interferon alpha-1/13, ScFv antibody | | Authors: | Ouyang, S, Zhao, L.X, Liang, W, Shaw, N, Liu, Z.-J, Liang, M.-F. | | Deposit date: | 2011-12-04 | | Release date: | 2012-02-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights into a human anti-IFN antibody exerting therapeutic potential for systemic lupus erythematosus
J.Mol.Med., 2012
|
|
3B6Y
 
 | | Crystal Structure of the Second HIN-200 Domain of Interferon-Inducible Protein 16 | | Descriptor: | Gamma-interferon-inducible protein Ifi-16, SULFATE ION | | Authors: | Liao, J.C.C, Lam, R, Ravichandran, M, Duan, S, Tempel, W, Chirgadze, N.Y, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-10-29 | | Release date: | 2007-11-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure Analysis of the Second HIN Domain of IFI16.
To be Published
|
|
4D5N
 
 | | Cryo-EM structures of ribosomal 80S complexes with termination factors and cricket paralysis virus IRES reveal the IRES in the translocated state | | Descriptor: | CRICKET PARALYSIS VIRUS IRES RNA, EUKARYOTIC PEPTIDE CHAIN RELEASE FACTOR SUBUNIT 1 | | Authors: | Muhs, M, Hilal, T, Mielke, T, Skabkin, M.A, Sanbonmatsu, K.Y, Pestova, T.V, Spahn, C.M.T. | | Deposit date: | 2014-11-06 | | Release date: | 2015-02-04 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Cryo-Em of Ribosomal 80S Complexes with Termination Factors Reveals the Translocated Cricket Paralysis Virus Ires.
Mol.Cell, 57, 2015
|
|
4QGU
 
 | | protein domain complex with ssDNA | | Descriptor: | DNA (5'-D(P*AP*GP*GP*CP*CP*GP*GP*CP*GP*TP*GP*A)-3'), Gamma-interferon-inducible protein 16 | | Authors: | Ni, X, Ru, H, Zhao, L, Shaw, N, Ding, W, Songying, O, Liu, Z.-J. | | Deposit date: | 2014-05-25 | | Release date: | 2015-06-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.545 Å) | | Cite: | New insights into the structural basis of DNA recognition by HINa and HINb domains of IFI16.
J Mol Cell Biol, 8, 2016
|
|
3RLN
 
 | |
1G9O
 
 | | FIRST PDZ DOMAIN OF THE HUMAN NA+/H+ EXCHANGER REGULATORY FACTOR | | Descriptor: | NHE-RF | | Authors: | Karthikeyan, S, Leung, T, Birrane, G, Webster, G, Ladias, J.A.A. | | Deposit date: | 2000-11-26 | | Release date: | 2001-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the PDZ1 domain of human Na(+)/H(+) exchanger regulatory factor provides insights into the mechanism of carboxyl-terminal leucine recognition by class I PDZ domains.
J.Mol.Biol., 308, 2001
|
|
2D4H
 
 | | Crystal-structure of the N-terminal large GTPase Domain of human Guanylate Binding protein 1 (hGBP1) in complex with GMP | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, Interferon-induced guanylate-binding protein 1 | | Authors: | Ghosh, A, Praefcke, G.J.K, Renault, L, Wittinghofer, A, Herrmann, C. | | Deposit date: | 2005-10-19 | | Release date: | 2006-03-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | How guanylate-binding proteins achieve assembly-stimulated processive cleavage of GTP to GMP.
Nature, 440, 2006
|
|
1DG3
 
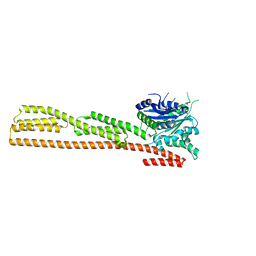 | | STRUCTURE OF HUMAN GUANYLATE BINDING PROTEIN-1 IN NUCLEOTIDE FREE FORM | | Descriptor: | PROTEIN (INTERFERON-INDUCED GUANYLATE-BINDING PROTEIN 1) | | Authors: | Prakash, B, Praefcke, G.J.K, Renault, L, Wittinghofer, A, Herrmann, C. | | Deposit date: | 1999-11-23 | | Release date: | 2000-10-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of human guanylate-binding protein 1 representing a unique class of GTP-binding proteins.
Nature, 403, 2000
|
|
2B8W
 
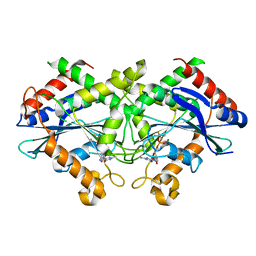 | | Crystal-structure of the N-terminal Large GTPase Domain of human Guanylate Binding protein 1 (hGBP1) in complex with GMP/AlF4 | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, Interferon-induced guanylate-binding protein 1, MAGNESIUM ION, ... | | Authors: | Ghosh, A, Praefcke, G.J.K, Renault, L, Wittinghofer, A, Herrmann, C. | | Deposit date: | 2005-10-10 | | Release date: | 2006-03-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | How guanylate-binding proteins achieve assembly-stimulated processive cleavage of GTP to GMP.
Nature, 440, 2006
|
|
2B92
 
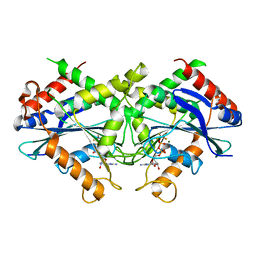 | | Crystal-structure of the N-terminal Large GTPase Domain of human Guanylate Binding protein 1 (hGBP1) in complex with GDP/AlF3 | | Descriptor: | ALUMINUM FLUORIDE, GUANOSINE-5'-DIPHOSPHATE, Interferon-induced guanylate-binding protein 1, ... | | Authors: | Ghosh, A, Praefcke, G.J.K, Renault, L, Wittinghofer, A, Herrmann, C. | | Deposit date: | 2005-10-10 | | Release date: | 2006-03-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | How guanylate-binding proteins achieve assembly-stimulated processive cleavage of GTP to GMP.
Nature, 440, 2006
|
|
1F5N
 
 | | HUMAN GUANYLATE BINDING PROTEIN-1 IN COMPLEX WITH THE GTP ANALOGUE, GMPPNP. | | Descriptor: | INTERFERON-INDUCED GUANYLATE-BINDING PROTEIN 1, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Prakash, B, Renault, L, Praefcke, G.J.K, Herrmann, C, Wittinghofer, A. | | Deposit date: | 2000-06-15 | | Release date: | 2000-09-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Triphosphate structure of guanylate-binding protein 1 and implications for nucleotide binding and GTPase mechanism.
EMBO J., 19, 2000
|
|
3QU3
 
 | | Crystal structure of IRF-7 DBD apo form | | Descriptor: | 1,2-ETHANEDIOL, Interferon regulatory factor 7, SODIUM ION | | Authors: | De Ioannes, P.E, Escalante, C.R, Aggarwal, A.K. | | Deposit date: | 2011-02-23 | | Release date: | 2011-06-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structures of apo IRF-3 and IRF-7 DNA binding domains: effect of loop L1 on DNA binding.
Nucleic Acids Res., 39, 2011
|
|
