4RVI
 
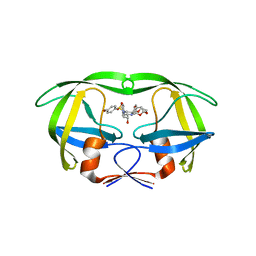 | | Crystal structure of multidrug-resistant clinical isolate A02 HIV-1 protease in complex with non-peptidic inhibitor, GRL0519 | | Descriptor: | (3R,3aS,3bR,6aS,7aS)-octahydrodifuro[2,3-b:3',2'-d]furan-3-yl [(1S,2R)-1-benzyl-2-hydroxy-3-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}propyl]carbamate, HIV-1 protease | | Authors: | Yedidi, R.S, Garimella, H, Kaufman, J.D, Das, D, Wingfield, P.T, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2014-11-26 | | Release date: | 2016-05-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Enhanced antiviral activity by the P2-tris-tetrahydrofuran moiety of GRL-0519, a novel nonpeptidic HIV-1 protease inhibitor (PI), against multi-PI-resistant and highly darunavir-resistant strains of HIV-1
To be Published
|
|
6ZYJ
 
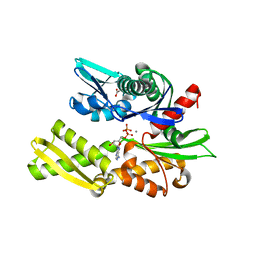 | | Crystal structure of Hsc70 ATPase domain in complex with ADP and calcium | | Descriptor: | ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, ... | | Authors: | Yan, Y, Preissler, S, Ron, D. | | Deposit date: | 2020-08-01 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Calcium depletion challenges endoplasmic reticulum proteostasis by destabilising BiP-substrate complexes.
Elife, 9, 2020
|
|
8BOV
 
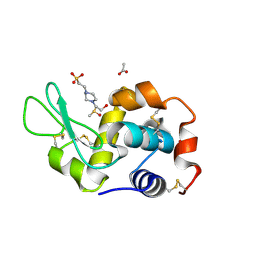 | | X-ray structure of the adduct formed upon reaction of the five-coordinate Pt(II) complex, 1-Me,Me, with HEWL at pH 7.5 | | Descriptor: | 1-[1,3-dimethyl-4-(1~{H}-1,2,3-triazol-5-yl)imidazol-1-ium-2-yl]-1,2',11'-trimethyl-spiro[1$l^{6}-platinacycloprop-2-ene-1,15'-1,12-diaza-15$l^{6}-platinatetracyclo[10.2.1.0^{5,14}.0^{8,13}]pentadeca-2,4,6,8,10,13-hexaene], 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETATE ION, ... | | Authors: | Ferraro, G, Tito, G, Merlino, A. | | Deposit date: | 2022-11-15 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Impact of Hydrophobic Chains in Five-Coordinate Glucoconjugate Pt(II) Anticancer Agents.
Int J Mol Sci, 24, 2023
|
|
8BOY
 
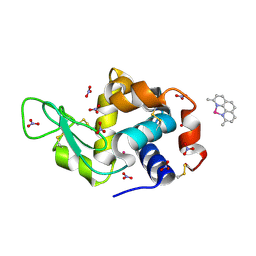 | | X-ray structure of the adduct formed upon reaction of the five-coordinate Pt(II) complex, 1-Me,Me, with HEWL at pH 4.0 | | Descriptor: | 1-[1,3-dimethyl-4-(1~{H}-1,2,3-triazol-5-yl)imidazol-1-ium-2-yl]-1,2',11'-trimethyl-spiro[1$l^{6}-platinacycloprop-2-ene-1,15'-1,12-diaza-15$l^{6}-platinatetracyclo[10.2.1.0^{5,14}.0^{8,13}]pentadeca-2,4,6,8,10,13-hexaene], Lysozyme C, NITRATE ION, ... | | Authors: | Ferraro, G, Tito, G, Merlino, A. | | Deposit date: | 2022-11-15 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Impact of Hydrophobic Chains in Five-Coordinate Glucoconjugate Pt(II) Anticancer Agents.
Int J Mol Sci, 24, 2023
|
|
8R7G
 
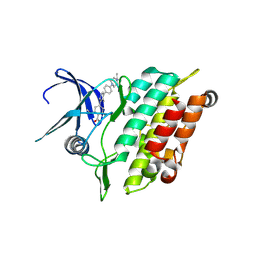 | | Crystal structure of the kinase domain of ACVR1 (ALK2) with M4K2234 | | Descriptor: | 2-fluoranyl-6-methoxy-4-[4-methyl-5-[4-(4-propan-2-ylpiperazin-1-yl)phenyl]pyridin-3-yl]benzamide, Activin receptor type I | | Authors: | Williams, E.P, Cros, J, Ensan, D, Smil, D, Edwards, A.M, O'Meara, J.A, Fernandez-Cid, A, Isaac, M.B, Al-awar, R, Bullock, A.N. | | Deposit date: | 2023-11-24 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Targeting ALK2: An Open Science Approach to Developing Therapeutics for the Treatment of Diffuse Intrinsic Pontine Glioma.
J.Med.Chem., 63, 2020
|
|
6V5L
 
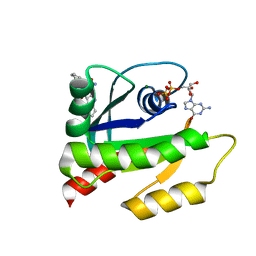 | | The HADDOCK structure model of GDP KRas in complex with its allosteric inhibitor E22 | | Descriptor: | (2R)-2-[2-(1H-indole-3-carbonyl)hydrazinyl]-2-phenylacetamide, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Wang, X, Gupta, A.K, Prakash, P, Putkey, J.P, Gorfe, A.A. | | Deposit date: | 2019-12-04 | | Release date: | 2019-12-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Multi target ensemble based virtual screening yields novel allosteric KRAS inhibitors at high success rate
Chemical Biology & Drug Design, 94, 2019
|
|
6ION
 
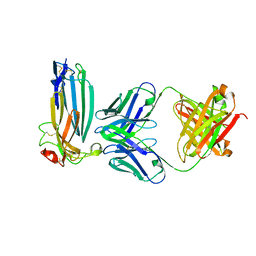 | | The complex of C4.4A with its antibody 11H10 Fab fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ly6/PLAUR domain-containing protein 3, ... | | Authors: | Huang, M.D, Jiang, Y.B, Yuan, C, Lin, L. | | Deposit date: | 2018-10-30 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal Structures of Human C4.4A Reveal the Unique Association of Ly6/uPAR/alpha-neurotoxin Domain
Int J Biol Sci, 16, 2020
|
|
4RYC
 
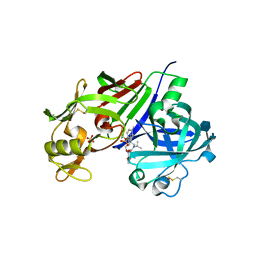 | |
8R4V
 
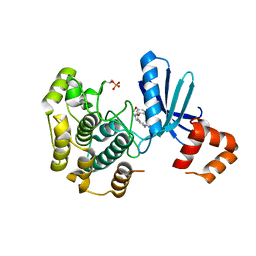 | | Structure of Salt-inducible kinase 3 in complex with inhibitor | | Descriptor: | 1-(2,4-dimethoxyphenyl)-3-(2,6-dimethylphenyl)-1-[6-[[4-(4-methylpiperazin-1-yl)phenyl]amino]pyrimidin-4-yl]urea, Serine/threonine-protein kinase SIK3 | | Authors: | Kack, H, Oster, L. | | Deposit date: | 2023-11-14 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structures of salt-inducible kinase 3 in complex with inhibitors reveal determinants for binding and selectivity.
J.Biol.Chem., 300, 2024
|
|
4S30
 
 | |
6ZYH
 
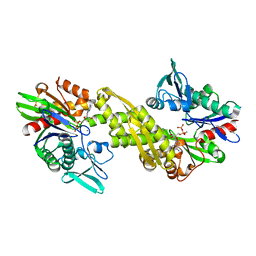 | |
4RZ1
 
 | |
6VIT
 
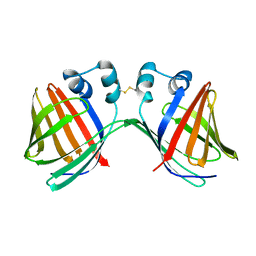 | |
8SF8
 
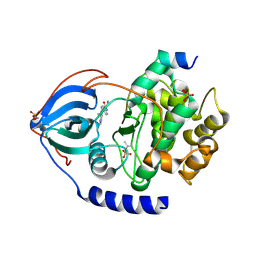 | | Structure of bovine PKA bound to (R)-N-(4-(1H-pyrrolo[2,3-b]pyridin-4-yl)phenyl)-2-amino-4-methylpentanamide | | Descriptor: | N-[4-(1H-pyrrolo[2,3-b]pyridin-4-yl)phenyl]-D-leucinamide, cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase inhibitor alpha | | Authors: | Coker, J.A, Arya, T, Goins, C.M, Maw, J.J, Macdonald, J.D, Stauffer, S.R. | | Deposit date: | 2023-04-10 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery and Characterization of Selective, First-in-Class Inhibitors of Citron Kinase.
J.Med.Chem., 67, 2024
|
|
5FBI
 
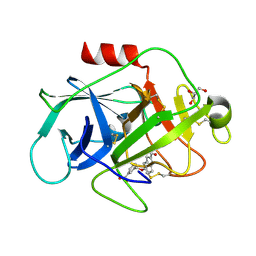 | | COMPLEMENT FACTOR D IN COMPLEX WITH COMPOUND 3b | | Descriptor: | 3-[(2-aminocarbonyl-1~{H}-indol-5-yl)oxymethyl]benzoic acid, Complement factor D, GLYCEROL | | Authors: | Ostermann, N, Zink, F. | | Deposit date: | 2015-12-14 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Small-molecule factor D inhibitors targeting the alternative complement pathway.
Nat.Chem.Biol., 12, 2016
|
|
7A49
 
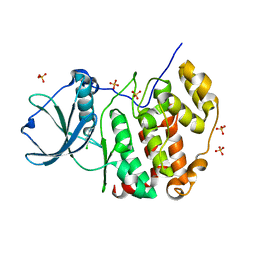 | | Crystal structure of human protein kinase CK2alpha (CSNK2A1 gene product) in complex with the ATP-competitive inhibitor 6-bromo-5-chloro-1H-triazolo[4,5-b]pyridine | | Descriptor: | 6-bromanyl-5-chloranyl-1~{H}-[1,2,3]triazolo[4,5-b]pyridine, Casein kinase II subunit alpha, SULFATE ION | | Authors: | Niefind, K, Lindenblatt, D, Toelzer, C, Bretner, M, Chojnacki, K, Wielechowska, M, Winska, P. | | Deposit date: | 2020-08-19 | | Release date: | 2020-12-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Synthesis, biological properties and structural study of new halogenated azolo[4,5-b]pyridines as inhibitors of CK2 kinase.
Bioorg.Chem., 106, 2021
|
|
6ISE
 
 | | Crystal structure of AMPPNP bound CK2 alpha from C. neoformans | | Descriptor: | 1,2-ETHANEDIOL, Casein kinase II subunit alpha, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Cho, H.S. | | Deposit date: | 2018-11-16 | | Release date: | 2019-11-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of AMPPNP bound CK2 alpha from C. neoformans
To be published
|
|
8RPM
 
 | |
8S2V
 
 | |
6VPG
 
 | |
4TN6
 
 | | CK1d in complex with inhibitor | | Descriptor: | 4-{4-(4-fluorophenyl)-1-[1-(1,2-oxazol-3-ylmethyl)piperidin-4-yl]-1H-imidazol-5-yl}pyrimidin-2-amine, Casein kinase I isoform delta, SULFATE ION | | Authors: | Liu, S. | | Deposit date: | 2014-06-03 | | Release date: | 2015-08-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | CK1D IN COMPLEX WITH inhibitor
To Be Published
|
|
5F8Z
 
 | | The crystal structure of human Plasma Kallikrein in complex with its peptide inhibitor pkalin-1 | | Descriptor: | CYS-PRO-ALA-ARG-PHE-M70-ALA-LEU-PHE-CYS, Plasma kallikrein LIGHT CHAIN, SULFATE ION, ... | | Authors: | Xu, M, Jiang, L, Xu, P, Luo, Z, Andreasen, P, Huang, M. | | Deposit date: | 2015-12-09 | | Release date: | 2016-12-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The crystal structure of human Plasma Kallikrein in complex with its peptide inhibitor pkalin-1
To Be Published
|
|
8BNQ
 
 | |
6VPM
 
 | |
8S2U
 
 | |
