3LLM
 
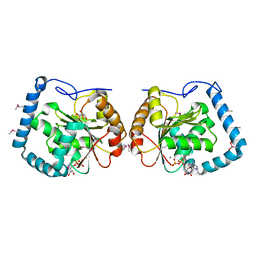 | | Crystal Structure Analysis of a RNA Helicase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent RNA helicase A, CACODYLATE ION, ... | | Authors: | Schutz, P, Karlberg, T, Collins, R, Arrowsmith, C.H, Berglund, H, Bountra, C, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Kallas, A, Kraulis, P, Kotenyova, T, Kotzsch, A, Markova, N, Moche, M, Nielsen, T.K, Nordlund, P, Nyman, T, Persson, C, Roos, A.K, Siponen, M.I, Svensson, L, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Wahlberg, E, Weigelt, J, Welin, M, Wisniewska, M, Schuler, H.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-01-29 | | Release date: | 2010-05-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of human RNA helicase A (DHX9): structural basis for unselective nucleotide base binding in a DEAD-box variant protein.
J.Mol.Biol., 400, 2010
|
|
5ZM0
 
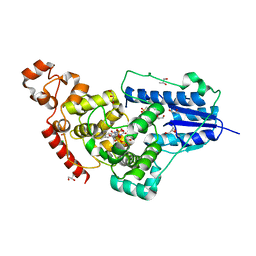 | | X-ray structure of animal-like Cryptochrome from Chlamydomonas reinhardtii | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, Cryptochrome photoreceptor, ... | | Authors: | Franz, S, Ignatz, E, Wenzel, S, Zielosko, H, Gusti Ngurah Putu, E.P, Maestre-Reyna, M, Tsai, M.-D, Yamamoto, J, Mittag, M, Essen, L.-O. | | Deposit date: | 2018-03-31 | | Release date: | 2018-07-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the bifunctional cryptochrome aCRY from Chlamydomonas reinhardtii
Nucleic Acids Res., 46, 2018
|
|
1YTA
 
 | |
3ERK
 
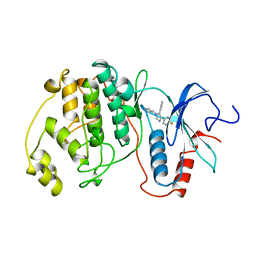 | | THE COMPLEX STRUCTURE OF THE MAP KINASE ERK2/SB220025 | | Descriptor: | 4-(4-FLUOROPHENYL)-1-(4-PIPERIDINYL)-5-(2-AMINO-4-PYRIMIDINYL)-IMIDAZOLE, EXTRACELLULAR REGULATED KINASE 2 | | Authors: | Wang, Z, Canagarajah, B, Boehm, J.C, Cobb, M.H, Young, P.R, Abdel-Meguid, S, Adams, J.L, Goldsmith, E.J. | | Deposit date: | 1998-07-09 | | Release date: | 1999-07-22 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of inhibitor selectivity in MAP kinases.
Structure, 6, 1998
|
|
3L3L
 
 | | PARP complexed with A906894 | | Descriptor: | 3-oxo-2-piperidin-4-yl-2,3-dihydro-1H-isoindole-4-carboxamide, Poly [ADP-ribose] polymerase 1 | | Authors: | Park, C.H. | | Deposit date: | 2009-12-17 | | Release date: | 2010-12-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery and SAR of substituted 3-oxoisoindoline-4-carboxamides as potent inhibitors of poly(ADP-ribose) polymerase (PARP) for the treatment of cancer.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3UR4
 
 | | Crystal structure of human WD repeat domain 5 with compound | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, SULFATE ION, ... | | Authors: | Dong, A, Dombrovski, L, Senisterra, G, Wernimont, A, Wasney, G.A, Allali Hassani, A, Nguyen, K.T, Smil, D, Bolshan, Y, Hajian, T, Poda, G, Chau, I, Al-Awar, R, Bountra, C, Weigelt, J, Edwards, A.M, Arrowsmith, C.H, Brown, P, Schapira, M, Vedadi, M, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-11-21 | | Release date: | 2011-12-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Small-molecule inhibition of MLL activity by disruption of its interaction with WDR5.
Biochem. J., 449, 2013
|
|
4FHI
 
 | | Development of synthetically accessible non-secosteroidal hybrid molecules combining vitamin D receptor agonism and histone deacetylase inhibition | | Descriptor: | N-hydroxy-2-{4-[3-(4-{[(2R)-2-hydroxy-3,3-dimethylbutyl]oxy}-3-methylphenyl)pentan-3-yl]-2-methylphenoxy}acetamide, SRC-1, Vitamin Nuclear Receptor | | Authors: | Fischer, J, Wang, T.T, Kaldre, D, Rochel, N, Moras, D, White, J.H, Gleason, J.L. | | Deposit date: | 2012-06-06 | | Release date: | 2012-09-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Synthetically accessible non-secosteroidal hybrid molecules combining vitamin d receptor agonism and histone deacetylase inhibition.
Chem.Biol., 19, 2012
|
|
2P1T
 
 | | Crystal structure of the ligand binding domain of the retinoid X receptor alpha in complex with 3-(2'-methoxy)-tetrahydronaphtyl cinnamic acid and a fragment of the coactivator TIF-2 | | Descriptor: | (2E)-3-[4-HYDROXY-3-(3-METHOXY-5,5,8,8-TETRAMETHYL-5,6,7,8-TETRAHYDRONAPHTHALEN-2-YL)PHENYL]ACRYLIC ACID, Nuclear receptor coactivator 2 peptide, Retinoic acid receptor RXR-alpha | | Authors: | Bourguet, W, Nahoum, V. | | Deposit date: | 2007-03-06 | | Release date: | 2007-10-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Modulators of the structural dynamics of the retinoid X receptor to reveal receptor function.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2P1V
 
 | | Crystal structure of the ligand binding domain of the retinoid X receptor alpha in complex with 3-(2'-propoxy)-tetrahydronaphtyl cinnamic acid and a fragment of the coactivator TIF-2 | | Descriptor: | (2E)-3-[4-HYDROXY-3-(5,5,8,8-TETRAMETHYL-3-PROPOXY-5,6,7,8-TETRAHYDRONAPHTHALEN-2-YL)PHENYL]ACRYLIC ACID, Nuclear receptor coactivator 2 peptide, Retinoic acid receptor RXR-alpha | | Authors: | Bourguet, W, Nahoum, V. | | Deposit date: | 2007-03-06 | | Release date: | 2007-10-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Modulators of the structural dynamics of the retinoid X receptor to reveal receptor function.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2DQR
 
 | |
1JIZ
 
 | | Crystal Structure Analysis of human Macrophage Elastase MMP-12 | | Descriptor: | CALCIUM ION, N-HYDROXY-2(R)-[[(4-METHOXYPHENYL)SULFONYL](3-PICOLYL)AMINO]-3-METHYLBUTANAMIDE HYDROCHLORIDE, ZINC ION, ... | | Authors: | Nar, H, Werle, K, Bauer, M.M.T, Dollinger, H, Jung, B. | | Deposit date: | 2001-07-03 | | Release date: | 2002-07-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of human macrophage elastase (MMP-12) in complex with a hydroxamic acid inhibitor.
J.Mol.Biol., 312, 2001
|
|
1JBR
 
 | | Crystal Structure of the Ribotoxin Restrictocin and a 31-mer SRD RNA Inhibitor | | Descriptor: | 31-mer SRD RNA analog, 5'-R(*GP*CP*GP*CP*UP*CP*CP*UP*CP*AP*GP*UP*AP*CP*GP*AP*GP*(A23))-3', 5'-R(*GP*GP*AP*AP*CP*CP*GP*GP*AP*GP*CP*GP*C)-3', ... | | Authors: | Yang, X, Gerczei, T, Glover, L, Correll, C.C. | | Deposit date: | 2001-06-06 | | Release date: | 2001-10-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structures of restrictocin-inhibitor complexes with implications for RNA recognition and base flipping.
Nat.Struct.Biol., 8, 2001
|
|
3IMJ
 
 | |
3IQR
 
 | |
1SJS
 
 | |
3IBW
 
 | | Crystal Structure of the ACT domain from GTP pyrophosphokinase of Chlorobium tepidum. Northeast Structural Genomics Consortium Target CtR148A | | Descriptor: | GTP pyrophosphokinase | | Authors: | Vorobiev, S, Su, M, Seetharaman, J, Janjua, J, Xiao, R, Ciccosanti, C, Wang, H, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-07-17 | | Release date: | 2009-07-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal Structure of the ACT domain from GTP pyrophosphokinase of Chlorobium tepidum.
To be Published
|
|
4CLJ
 
 | | Structure of L1196M Mutant Human Anaplastic Lymphoma Kinase in Complex with PF-06463922 ((10R)-7-amino-12-fluoro-2,10,16-trimethyl- 15-oxo-10,15,16,17-tetrahydro-2H-8,4-(metheno)pyrazolo(4,3-h)(2,5,11) benzoxadiazacyclotetradecine-3-carbonitrile). | | Descriptor: | (10R)-7-amino-12-fluoro-2,10,16-trimethyl-15-oxo-10,15,16,17-tetrahydro-2H-8,4-(metheno)pyrazolo[4,3-h][2,5,11]benzoxadiazacyclotetradecine-3-carbonitrile, ALK TYROSINE KINASE RECEPTOR | | Authors: | McTigue, M.A, Deng, Y.L, Liu, W, Brooun, A, Stewart, A.E. | | Deposit date: | 2014-01-14 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Discovery of (10R)-7-Amino-12-Fluoro-2,10,16-Trimethyl-15-Oxo-10,15,16,17-Tetrahydro-2H-8,4-(Metheno)Pyrazolo[4,3-H][2,5,11]Benzoxadiazacyclotetradecine-3-Carbonitrile (Pf-06463922), a Macrocyclic Inhibitor of Alk/Ros1 with Pre-Clinical Brain Exposure and Broad Spectrum Potency Against Alk-Resistant Mutations.
J.Med.Chem., 57, 2014
|
|
2LY9
 
 | | Solution NMR Structure of Homeobox 2 Domain from Human ZHX1 repressor, Northeast Structural Genomics Consortium (NESG) Target HR7907F | | Descriptor: | Zinc fingers and homeoboxes protein 1 | | Authors: | Xu, X, Eletsky, A, Mills, J.L, Pulavarti, S.V.S.R.K, Lee, D, Kohan, E, Janjua, H, Xiao, R, Acton, T.B, Everett, J.K, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-09-14 | | Release date: | 2012-11-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of Homeobox 2 Domain from Human ZHX1 repressor, Northeast Structural Genomics Consortium (NESG) Target HR7907F
To be Published
|
|
1T7C
 
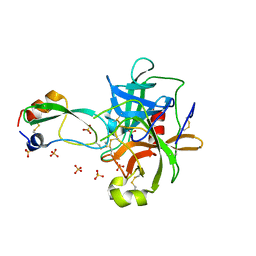 | | CRYSTAL STRUCTURE OF THE P1 GLU BPTI MUTANT- BOVINE CHYMOTRYPSIN COMPLEX | | Descriptor: | Chymotrypsin A, Pancreatic trypsin inhibitor, SULFATE ION | | Authors: | Czapinska, H, Helland, R, Otlewski, J, Smalas, A.O. | | Deposit date: | 2004-05-09 | | Release date: | 2005-03-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structures of five bovine chymotrypsin complexes with P1 BPTI variants.
J.Mol.Biol., 344, 2004
|
|
1JH6
 
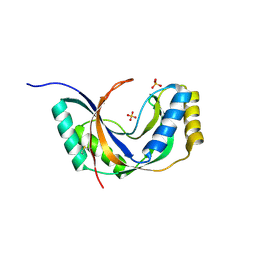 | | Semi-reduced Cyclic Nucleotide Phosphodiesterase from Arabidopsis thaliana | | Descriptor: | SULFATE ION, cyclic phosphodiesterase | | Authors: | Hofmann, A, Grella, M, Botos, I, Filipowicz, W, Wlodawer, A. | | Deposit date: | 2001-06-27 | | Release date: | 2002-02-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of the semireduced and inhibitor-bound forms of cyclic nucleotide phosphodiesterase from Arabidopsis thaliana.
J.Biol.Chem., 277, 2002
|
|
1JKH
 
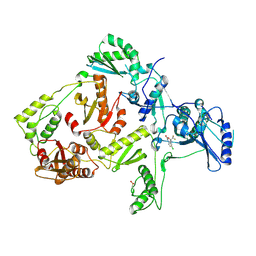 | | CRYSTAL STRUCTURE OF Y181C MUTANT HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH DMP-266(EFAVIRENZ) | | Descriptor: | (-)-6-CHLORO-4-CYCLOPROPYLETHYNYL-4-TRIFLUOROMETHYL-1,4-DIHYDRO-2H-3,1-BENZOXAZIN-2-ONE, HIV-1 RT, A-CHAIN, ... | | Authors: | Ren, J, Nichols, C, Bird, L, Chamberlain, P, Weaver, K, Short, S, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2001-07-12 | | Release date: | 2001-10-03 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural mechanisms of drug resistance for mutations at codons 181 and 188 in HIV-1 reverse transcriptase and the improved resilience of second generation non-nucleoside inhibitors.
J.Mol.Biol., 312, 2001
|
|
4CLI
 
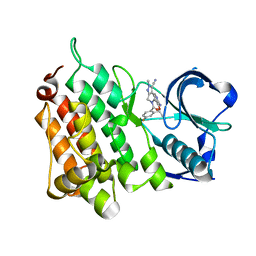 | | Structure of the Human Anaplastic Lymphoma Kinase in Complex with PF- 06463922 ((10R)-7-amino-12-fluoro-2,10,16-trimethyl-15-oxo-10,15,16, 17-tetrahydro-2H-8,4-(metheno)pyrazolo(4,3-h)(2,5,11) benzoxadiazacyclotetradecine-3-carbonitrile). | | Descriptor: | (10R)-7-amino-12-fluoro-2,10,16-trimethyl-15-oxo-10,15,16,17-tetrahydro-2H-8,4-(metheno)pyrazolo[4,3-h][2,5,11]benzoxadiazacyclotetradecine-3-carbonitrile, ALK TYROSINE KINASE RECEPTOR | | Authors: | McTigue, M.A, Deng, Y.L, Liu, W, Brooun, A, Stewart, A.E. | | Deposit date: | 2014-01-14 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery of (10R)-7-Amino-12-Fluoro-2,10,16-Trimethyl-15-Oxo-10,15,16,17-Tetrahydro-2H-8,4-(Metheno)Pyrazolo[4,3-H][2,5,11]Benzoxadiazacyclotetradecine-3-Carbonitrile (Pf-06463922), a Macrocyclic Inhibitor of Alk/Ros1 with Pre-Clinical Brain Exposure and Broad Spectrum Potency Against Alk-Resistant Mutations.
J.Med.Chem., 57, 2014
|
|
1JLG
 
 | | CRYSTAL STRUCTURE OF Y188C MUTANT HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH UC-781 | | Descriptor: | 2-METHYL-FURAN-3-CARBOTHIOIC ACID [4-CHLORO-3-(3-METHYL-BUT-2-ENYLOXY)-PHENYL]-AMIDE, HIV-1 RT A-chain, HIV-1 RT B-chain | | Authors: | Ren, J, Nichols, C, Bird, L, Chamberlain, P, Weaver, K, Short, S, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2001-07-16 | | Release date: | 2001-10-03 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural mechanisms of drug resistance for mutations at codons 181 and 188 in HIV-1 reverse transcriptase and the improved resilience of second generation non-nucleoside inhibitors.
J.Mol.Biol., 312, 2001
|
|
1TKX
 
 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH GW490745 | | Descriptor: | 4-[(CYCLOPROPYLETHYNYL)OXY]-6-FLUORO-3-ISOPROPYLQUINOLIN-2(1H)-ONE, Pol polyprotein, Reverse transcriptase, ... | | Authors: | Ren, J, Hopkins, A.L, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2004-06-09 | | Release date: | 2004-12-07 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Design of non-nucleoside inhibitors of HIV-1 reverse transcriptase with improved drug resistance properties. 2.
J.Med.Chem., 47, 2004
|
|
3IQN
 
 | |
