3M1S
 
 | | Structure of Ruthenium Half-Sandwich Complex Bound to Glycogen Synthase Kinase 3 | | Descriptor: | Glycogen synthase kinase-3 beta, Ruthenium pyridocarbazole | | Authors: | Atilla-Gokcumen, G.E, Di Costanzo, L, Zimmermann, G, Meggers, E. | | Deposit date: | 2010-03-05 | | Release date: | 2010-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.134 Å) | | Cite: | Structure of anticancer ruthenium half-sandwich complex bound to glycogen synthase kinase 3beta
J.Biol.Inorg.Chem., 16, 2011
|
|
1SRA
 
 | | STRUCTURE OF A NOVEL EXTRACELLULAR CA2+-BINDING MODULE IN BM-40(SLASH)SPARC(SLASH)OSTEONECTIN | | Descriptor: | CALCIUM ION, SPARC | | Authors: | Hohenester, E, Maurer, P, Hohenadl, C, Timpl, R, Jansonius, J.N, Engel, J. | | Deposit date: | 1995-08-21 | | Release date: | 1996-03-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a novel extracellular Ca(2+)-binding module in BM-40.
Nat.Struct.Biol., 3, 1996
|
|
8R6O
 
 | | Tubulin-4AZA2996 complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, Detyrosinated tubulin alpha-1B chain, ... | | Authors: | Herman, J, Vanstreels, E, Bardiot, D, Prota, A.E, Olieric, N, Gao, L.-J, Vercruysse, T, Daems, T, Waer, M, Herdewijn, P, Louat, T, Steinmetz, M.O, De Jonghe, S, Sprangers, B, Daelemans, D. | | Deposit date: | 2023-11-22 | | Release date: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | 3-nitropyridine analogues as novel microtubule-targeting agents.
Plos One, 19, 2024
|
|
3PF3
 
 | | Crystal structure of a mutant (C202A) of Triosephosphate isomerase from Giardia lamblia derivatized with MMTS | | Descriptor: | CALCIUM ION, GLYCEROL, SULFATE ION, ... | | Authors: | Enriquez-Flores, S, Rodriguez-Romero, A, Hernandez-Santoyo, A, Reyes-Vivas, H. | | Deposit date: | 2010-10-27 | | Release date: | 2011-06-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.098 Å) | | Cite: | Determining the molecular mechanism of inactivation by chemical modification of triosephosphate isomerase from the human parasite Giardia lamblia: A study for antiparasitic drug design.
Proteins, 79, 2011
|
|
8AEV
 
 | | Human acetylcholinesterase in complex with N,N,N-trimethyl-2-oxo-2-(2-(pyridin-2-ylmethylene)hydrazineyl)ethan-1-aminium | | Descriptor: | 1-(2-(2-((6-(dihydroxymethyl)-2-phenylpyrimidin-4-yl)methylene)hydrazineyl)-2-oxoethyl)pyridin-1-ium, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Nachon, F, Dias, J, Brazzolotto, X. | | Deposit date: | 2022-07-13 | | Release date: | 2023-06-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Grid-Type Quaternary Metallosupramolecular Compounds Inhibit Human Cholinesterases through Dynamic Multivalent Interactions.
Chembiochem, 23, 2022
|
|
8VLB
 
 | | Crystal structure of EloBC-VHL-CDO1 complex bound to compound 4 molecular glue | | Descriptor: | CITRIC ACID, Cysteine dioxygenase type 1, Elongin-B, ... | | Authors: | Shu, W, Ma, X, Tutter, A, Buckley, D, Golosov, A, Michaud, G. | | Deposit date: | 2024-01-11 | | Release date: | 2024-03-13 | | Last modified: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A small-molecule VHL molecular glue degrader for cysteine dioxygenase 1.
Nat.Chem.Biol., 2025
|
|
8UZH
 
 | |
8P1Z
 
 | | Arabidopsis thaliana cytosolic seryl-tRNA synthetase in addition of dithiothreitol (DTT) | | Descriptor: | Serine--tRNA ligase, cytoplasmic | | Authors: | Kekez, I, Soic, R, Rokov-Plavec, J, Matkovic-Calogovic, D. | | Deposit date: | 2023-05-13 | | Release date: | 2024-05-22 | | Last modified: | 2024-12-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Evolutionarily conserved cysteines in plant cytosolic seryl-tRNA synthetase are important for its resistance to oxidation.
Febs Lett., 597, 2023
|
|
6R35
 
 | | Structure of the LecB lectin from Pseudomonas aeruginosa strain PAO1 in complex with lewis x tetrasaccharide | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Fucose-binding lectin PA-IIL, ... | | Authors: | Lepsik, M, Sommer, R, Kuhaudomlarp, S, Lelimousin, M, Varrot, A, Titz, A, Imberty, A. | | Deposit date: | 2019-03-19 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Induction of rare conformation of oligosaccharide by binding to calcium-dependent bacterial lectin: X-ray crystallography and modelling study.
Eur.J.Med.Chem., 177, 2019
|
|
8P1Y
 
 | | Arabidopsis thaliana mutated variant C244S of seryl-tRNA synthetase | | Descriptor: | Serine--tRNA ligase, cytoplasmic | | Authors: | Kekez, I, Soic, R, Rokov-Plavec, J, Matkovic-Calogovic, D. | | Deposit date: | 2023-05-13 | | Release date: | 2024-05-22 | | Last modified: | 2024-12-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Evolutionarily conserved cysteines in plant cytosolic seryl-tRNA synthetase are important for its resistance to oxidation.
Febs Lett., 597, 2023
|
|
6H1B
 
 | | Structure of amide bond synthetase Mcba K483A mutant from Marinactinospora thermotolerans | | Descriptor: | 1-ethanoyl-9~{H}-pyrido[3,4-b]indole-3-carboxylic acid, ADENOSINE MONOPHOSPHATE, Fatty acid CoA ligase | | Authors: | Rowlinson, B, Petchey, M, Cuetos, A, Frese, A, Dannevald, S, Grogan, G. | | Deposit date: | 2018-07-11 | | Release date: | 2018-09-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Broad Aryl Acid Specificity of the Amide Bond Synthetase McbA Suggests Potential for the Biocatalytic Synthesis of Amides.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
1B2J
 
 | | CLOSTRIDIUM PASTEURIANUM RUBREDOXIN G43A MUTANT | | Descriptor: | FE (III) ION, PROTEIN (RUBREDOXIN) | | Authors: | Maher, M.J, Guss, J.M, Wilce, M.C.J, Wedd, A.G. | | Deposit date: | 1998-11-27 | | Release date: | 1999-05-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Rubredoxin from Clostridium pasteurianum. Structures of G10A, G43A and G10VG43A mutant proteins. Mutation of conserved glycine 10 to valine causes the 9-10 peptide link to invert.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
4OQP
 
 | |
1B13
 
 | | CLOSTRIDIUM PASTEURIANUM RUBREDOXIN G10A MUTANT | | Descriptor: | FE (III) ION, PROTEIN (RUBREDOXIN) | | Authors: | Maher, M.J, Guss, J.M, Wilce, M.C.J, Wedd, A.G. | | Deposit date: | 1998-11-26 | | Release date: | 1999-05-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Rubredoxin from Clostridium pasteurianum. Structures of G10A, G43A and G10VG43A mutant proteins. Mutation of conserved glycine 10 to valine causes the 9-10 peptide link to invert.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1B2O
 
 | | CLOSTRIDIUM PASTEURIANUM RUBREDOXIN G10VG43A MUTANT | | Descriptor: | FE (III) ION, PROTEIN (RUBREDOXIN) | | Authors: | Maher, M.J, Guss, J.M, Wilce, M.C.J, Wedd, A.G. | | Deposit date: | 1998-11-30 | | Release date: | 1999-05-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Rubredoxin from Clostridium pasteurianum. Structures of G10A, G43A and G10VG43A mutant proteins. Mutation of conserved glycine 10 to valine causes the 9-10 peptide link to invert.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
3H5W
 
 | | Crystal structure of the GluR2-ATD in space group P212121 without solvent | | Descriptor: | Glutamate receptor 2 | | Authors: | Jin, R, Singh, S.K, Gu, S, Furukawa, H, Sobolevsky, A, Zhou, J, Jin, Y, Gouaux, E. | | Deposit date: | 2009-04-22 | | Release date: | 2009-06-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.686 Å) | | Cite: | Crystal structure and association behaviour of the GluR2 amino-terminal domain.
Embo J., 28, 2009
|
|
2HEV
 
 | | Crystal structure of the complex between OX40L and OX40 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Tumor necrosis factor ligand superfamily member 4, Tumor necrosis factor receptor superfamily member 4 | | Authors: | Hymowitz, S.G, Compaan, D.M. | | Deposit date: | 2006-06-22 | | Release date: | 2006-08-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | The Crystal Structure of the Costimulatory OX40-OX40L Complex.
Structure, 14, 2006
|
|
7N8K
 
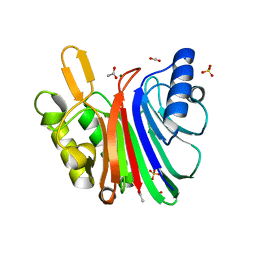 | | LINE-1 endonuclease domain complex with Mg | | Descriptor: | ACETATE ION, LINE-1 retrotransposable element ORF2 protein, MAGNESIUM ION, ... | | Authors: | Korolev, S, Miller, I. | | Deposit date: | 2021-06-15 | | Release date: | 2021-10-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural dissection of sequence recognition and catalytic mechanism of human LINE-1 endonuclease.
Nucleic Acids Res., 49, 2021
|
|
7N8S
 
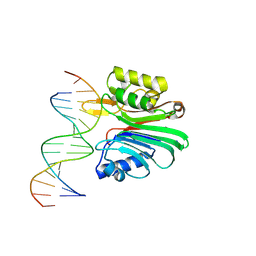 | | LINE-1 endonuclease domain complex with DNA | | Descriptor: | DNA (5'-D(*CP*CP*TP*TP*AP*AP*AP*AP*AP*GP*GP*AP*GP*CP*T)-3'), DNA (5'-D(*GP*CP*TP*CP*CP*TP*TP*TP*TP*TP*AP*AP*GP*GP*A)-3'), LINE-1 retrotransposable element ORF2 protein | | Authors: | Korolev, S, Miller, I. | | Deposit date: | 2021-06-15 | | Release date: | 2021-10-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structural dissection of sequence recognition and catalytic mechanism of human LINE-1 endonuclease.
Nucleic Acids Res., 49, 2021
|
|
2OHX
 
 | |
2BG9
 
 | |
6TM5
 
 | |
4BKE
 
 | | Recombinant human serum albumin with palmitic acid. Synthetic cationic antimicrobial peptides bind with their hydrophobic parts to drug site II of human serum albumin | | Descriptor: | PALMITIC ACID, SERUM ALBUMIN | | Authors: | Sivertsen, A, Isaksson, J, Leiros, H.-K.S, Svenson, J, Svendsen, J.-S, Brandsdal, B.-O. | | Deposit date: | 2013-04-24 | | Release date: | 2014-02-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Synthetic Cationic Antimicrobial Peptides Bind with Their Hydrophobic Parts to Drug Site II of Human Serum Albumin.
Bmc Struct.Biol., 14, 2014
|
|
8WZU
 
 | |
1XJO
 
 | | STRUCTURE OF AMINOPEPTIDASE | | Descriptor: | AMINOPEPTIDASE, CALCIUM ION, PHOSPHATE ION, ... | | Authors: | Greenblatt, H.M, Barra, D, Blumberg, S, Shoham, G. | | Deposit date: | 1996-10-09 | | Release date: | 1997-04-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Streptomyces griseus aminopeptidase: X-ray crystallographic structure at 1.75 A resolution.
J.Mol.Biol., 265, 1997
|
|
