8HAF
 
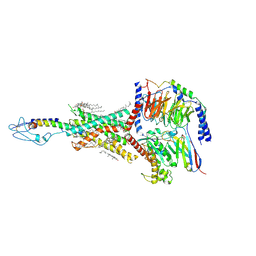 | | PTHrP-PTH1R-Gs complex | | Descriptor: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhao, L, Xu, H.E, Yuan, Q. | | Deposit date: | 2022-10-26 | | Release date: | 2022-12-21 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Molecular recognition of two endogenous hormones by the human parathyroid hormone receptor-1.
Acta Pharmacol.Sin., 44, 2023
|
|
8HA0
 
 | | Molecular recognition of two endogenous hormones by the human parathyroid hormone receptor-1 | | Descriptor: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhao, L, Xu, H.E, Yuan, Q. | | Deposit date: | 2022-10-26 | | Release date: | 2022-12-21 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.62 Å) | | Cite: | Molecular recognition of two endogenous hormones by the human parathyroid hormone receptor-1.
Acta Pharmacol.Sin., 44, 2023
|
|
8EIZ
 
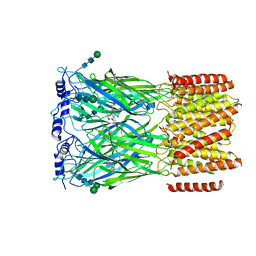 | | Cryo-EM structure of squid sensory receptor CRB1 | | Descriptor: | N-benzyl-2-(2,6-dimethylanilino)-N,N-diethyl-2-oxoethan-1-aminium, Squid sensory receptor CRB1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Kang, G, Kim, J.J, Allard, C.A.H, Valencia-Montoya, W.A, van Giesen, L, Kilian, P.B, Bai, X, Bellono, N.W, Hibbs, R.E. | | Deposit date: | 2022-09-15 | | Release date: | 2023-04-12 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Sensory specializations drive octopus and squid behaviour.
Nature, 616, 2023
|
|
6ZG7
 
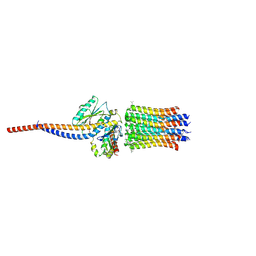 | | bovine ATP synthase rotor domain, state 1 | | Descriptor: | ATP synthase F(0) complex subunit C2, mitochondrial, ATP synthase subunit delta, ... | | Authors: | Spikes, T, Montgomery, M.G, Walker, J.E. | | Deposit date: | 2020-06-18 | | Release date: | 2020-09-09 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Structure of the dimeric ATP synthase from bovine mitochondria.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
9GL9
 
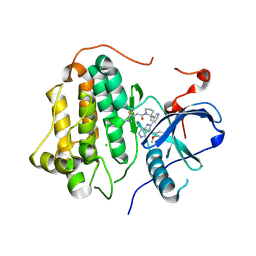 | | Wild-type EGFR bound with STX-721 | | Descriptor: | (2R,3S)-3-[(3-chloranyl-2-methoxy-phenyl)amino]-2-[3-[2-[(2R)-1-[(E)-4-(dimethylamino)but-2-enoyl]-2-methyl-pyrrolidin-2-yl]ethynyl]pyridin-4-yl]-1,2,3,5,6,7-hexahydropyrrolo[3,2-c]pyridin-4-one, CHLORIDE ION, Epidermal growth factor receptor | | Authors: | Hilbert, B.J, Brooijmans, N, Milgram, B.C, Pagliarini, R.A. | | Deposit date: | 2024-08-27 | | Release date: | 2025-05-14 | | Last modified: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (2.147 Å) | | Cite: | STX-721, a Covalent EGFR/HER2 Exon 20 Inhibitor, Utilizes Exon 20-Mutant Dynamic Protein States and Achieves Unique Mutant Selectivity Across Human Cancer Models.
Clin.Cancer Res., 2025
|
|
7TV0
 
 | |
7TUQ
 
 | |
6ZIK
 
 | | bovine ATP synthase rotor domain, state 3 | | Descriptor: | ATP synthase F(0) complex subunit C2, mitochondrial, ATP synthase subunit delta, ... | | Authors: | Spikes, T, Montgomery, M.G, Walker, J.E. | | Deposit date: | 2020-06-26 | | Release date: | 2020-09-09 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | Structure of the dimeric ATP synthase from bovine mitochondria.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6ZG8
 
 | | bovine ATP synthase rotor domain state 2 | | Descriptor: | ATP synthase F(0) complex subunit C2, mitochondrial, ATP synthase subunit delta, ... | | Authors: | Spikes, T, Montgomery, M.G, Walker, J.E. | | Deposit date: | 2020-06-18 | | Release date: | 2020-09-09 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Structure of the dimeric ATP synthase from bovine mitochondria.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5IWM
 
 | | 2.5A structure of GSK945237 with S.aureus DNA gyrase and DNA. | | Descriptor: | (1R)-1-[(4-{[(6,7-dihydro[1,4]dioxino[2,3-c]pyridazin-3-yl)methyl]amino}piperidin-1-yl)methyl]-9-fluoro-1,2-dihydro-4H-pyrrolo[3,2,1-ij]quinolin-4-one, DNA (5'-D(*AP*GP*CP*CP*GP*TP*AP*GP*GP*TP*TP*CP*AP*CP*CP*GP*CP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*GP*CP*GP*GP*TP*GP*AP*AP*CP*CP*TP*AP*CP*GP*GP*CP*T)-3'), ... | | Authors: | Bax, B.D, Miles, T.J. | | Deposit date: | 2016-03-22 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Novel tricyclics (e.g., GSK945237) as potent inhibitors of bacterial type IIA topoisomerases.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
7R02
 
 | | Mus musculus acetylcholinesterase in complex with N-(3-(diethylamino)propyl)-4-methyl-3-nitrobenzamide | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-METHOXYETHOXY)ETHANOL, 2-[2-[2-[2-[2-[2-(2-methoxyethoxy)ethoxy]ethoxy]ethoxy]ethoxy]ethoxy]ethanol, ... | | Authors: | Forsgren, N, Lindgren, C, Edvinsson, L, Linusson, A, Ekstrom, F. | | Deposit date: | 2022-02-01 | | Release date: | 2022-04-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Broad-Spectrum Antidote Discovery by Untangling the Reactivation Mechanism of Nerve-Agent-Inhibited Acetylcholinesterase.
Chemistry, 28, 2022
|
|
8ACW
 
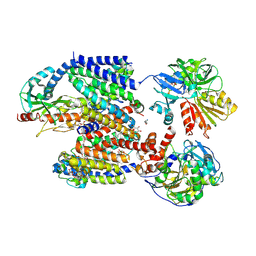 | | X-ray structure of Na+-NQR from Vibrio cholerae at 3.4 A resolution | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Fritz, G. | | Deposit date: | 2022-07-07 | | Release date: | 2023-07-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Conformational coupling of redox-driven Na + -translocation in Vibrio cholerae NADH:quinone oxidoreductase.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7BB5
 
 | |
7BGS
 
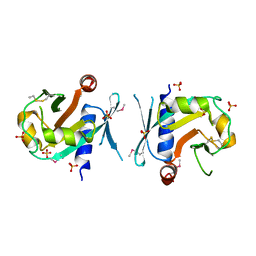 | | Archeal holliday junction resolvase from Thermus thermophilus phage 15-6 | | Descriptor: | Holliday junction resolvase, SULFATE ION | | Authors: | Hakansson, M, Ahlqvist, J, Linares Pasten, J.A, Jasilionis, A, Nordberg Karlsson, E, Al-Karadaghi, S. | | Deposit date: | 2021-01-08 | | Release date: | 2022-01-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure and initial characterization of a novel archaeal-like Holliday junction-resolving enzyme from Thermus thermophilus phage Tth15-6.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7BNX
 
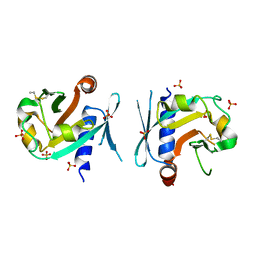 | | Archeal holliday junction resolvase from Thermus thermophilus phage 15-6 | | Descriptor: | Holliday junction resolvase, SULFATE ION | | Authors: | Hakansson, M, Ahlqvist, J, Linares Pasten, J.A, Jasilionis, A, Nordberg Karlsson, E, Al-Karadaghi, S. | | Deposit date: | 2021-01-22 | | Release date: | 2022-02-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.551 Å) | | Cite: | Crystal structure and initial characterization of a novel archaeal-like Holliday junction-resolving enzyme from Thermus thermophilus phage Tth15-6.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
8CO3
 
 | |
9H28
 
 | | Alternative conformation LGTV with TBEV prME | | Descriptor: | 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Genome polyprotein, ... | | Authors: | Bisikalo, K, Rosendal, E. | | Deposit date: | 2024-10-11 | | Release date: | 2024-11-06 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | The influence of the pre-membrane and envelope proteins on structure, pathogenicity and tropism of tick-borne encephalitis virus
Biorxiv, 2024
|
|
7TZK
 
 | | EPS8 SH3 Domain with NleH1 PxxDY Motif | | Descriptor: | Epidermal growth factor receptor kinase substrate 8, T3SS secreted effector NleH homolog | | Authors: | Grishin, A.M, Cygler, M. | | Deposit date: | 2022-02-15 | | Release date: | 2022-07-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Targeting of microvillus protein Eps8 by the NleH effector kinases from enteropathogenic E. coli
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
5YB0
 
 | |
4V4R
 
 | | Crystal structure of the whole ribosomal complex. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Petry, S, Brodersen, D.E, Murphy IV, F.V, Dunham, C.M, Selmer, M, Tarry, M.J, Kelley, A.C, Ramakrishnan, V. | | Deposit date: | 2005-09-30 | | Release date: | 2014-07-09 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (5.9 Å) | | Cite: | Crystal Structures of the Ribosome in Complex with Release Factors RF1 and RF2 Bound to a Cognate Stop Codon.
Cell(Cambridge,Mass.), 123, 2005
|
|
4V8J
 
 | | Crystal structure of the bacterial ribosome ram mutation G347U. | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Fagan, C.E, Dunkle, J.A, Maehigashi, T, Dunham, C.M. | | Deposit date: | 2011-12-20 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Reorganization of an intersubunit bridge induced by disparate 16S ribosomal ambiguity mutations mimics an EF-Tu-bound state.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4V4S
 
 | | Crystal structure of the whole ribosomal complex. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Petry, S, Brodersen, D.E, Murphy IV, F.V, Dunham, C.M, Selmer, M, Tarry, M.J, Kelley, A.C, Ramakrishnan, V. | | Deposit date: | 2005-10-12 | | Release date: | 2014-07-09 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (6.76 Å) | | Cite: | Crystal Structures of the Ribosome in Complex with Release Factors RF1 and RF2 Bound to a Cognate Stop Codon.
Cell(Cambridge,Mass.), 123, 2005
|
|
5HL4
 
 | | Acoustic injectors for drop-on-demand serial femtosecond crystallography | | Descriptor: | COBALT HEXAMMINE(III), FE (III) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Roessler, C.G, Agarwal, R, Allaire, M, Alonso-Mori, R, Andi, B, Bachega, J.F.R, Bommer, M, Brewster, A.S, Browne, M.C, Chatterjee, R, Cho, E, Cohen, A.E, Cowan, M, Datwani, S, Davidson, V.L, Defever, J, Eaton, B, Ellson, R, Feng, Y, Ghislain, L.P, Glownia, J.M, Han, G, Hattne, J, Hellmich, J, Heroux, A, Ibrahim, M, Kern, J, Kuczewski, A, Lemke, H.T, Liu, P, Majlof, L, McClintock, W.M, Myers, S, Nelsen, S, Olechno, J, Orville, A.M, Sauter, N.K, Soares, A.S, Soltis, M.S, Song, H, Stearns, R.G, Tran, R, Tsai, Y, Uervirojnangkoorn, M, Wilmot, C.M, Yachandra, V, Yano, J, Yukl, E.T, Zhu, D, Zouni, A. | | Deposit date: | 2016-01-14 | | Release date: | 2016-01-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Acoustic Injectors for Drop-On-Demand Serial Femtosecond Crystallography.
Structure, 24, 2016
|
|
1JKE
 
 | | D-Tyr tRNATyr deacylase from Escherichia coli | | Descriptor: | D-Tyr-tRNATyr deacylase, ZINC ION | | Authors: | Ferri-Fioni, M.L, Schmitt, E, Soutourina, J, Plateau, P, Mechulam, Y, Blanquet, S. | | Deposit date: | 2001-07-12 | | Release date: | 2002-01-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure of crystalline D-Tyr-tRNA(Tyr) deacylase. A representative of a new class of tRNA-dependent hydrolases.
J.Biol.Chem., 276, 2001
|
|
6PEG
 
 | | MIF with a allosteric inhibitor | | Descriptor: | 4-amino-5-hydroxy-6-[(E)-(3-{[3-(2-methylpropanoyl)pyrazolo[1,5-a]pyridin-2-yl]methyl}phenyl)diazenyl]naphthalene-1,3-disulfonic acid, ACETATE ION, GLYCEROL, ... | | Authors: | Asojo, O.A. | | Deposit date: | 2019-06-20 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inhibition of Macrophage Migration Inhibitory Factor by a Chimera of Two Allosteric Binders.
Acs Med.Chem.Lett., 11, 2020
|
|
