3M3E
 
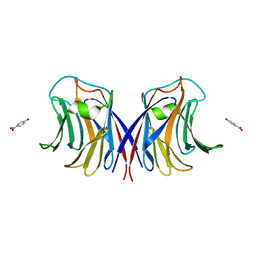 | |
6S9G
 
 | | Human Carbonic Anhydrase II in complex with fluorinated benzenesulfonamide | | Descriptor: | (4-CARBOXYPHENYL)(CHLORO)MERCURY, 2,3,5,6-tetrakis(fluoranyl)-4-propyl-benzenesulfonamide, Carbonic anhydrase 2, ... | | Authors: | Gloeckner, S, Ngo, K, Heine, A, Klebe, G. | | Deposit date: | 2019-07-12 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | The Influence of Varying Fluorination Patterns on the Thermodynamics and Kinetics of Benzenesulfonamide Binding to Human Carbonic Anhydrase II.
Biomolecules, 10, 2020
|
|
3RLL
 
 | | Crystal structure of the T877A androgen receptor ligand binding domain in complex with (S)-N-(4-Cyano-3-(trifluoromethyl)phenyl)-3-(4-cyanonaphthalen-1-yloxy)-2-hydroxy-2-methylpropanamide | | Descriptor: | (2S)-3-[(4-cyanonaphthalen-1-yl)oxy]-N-[4-cyano-3-(trifluoromethyl)phenyl]-2-hydroxy-2-methylpropanamide, Androgen receptor | | Authors: | Bohl, C.E, Duke, C.B, Jones, A, Dalton, J.T, Miller, D.D. | | Deposit date: | 2011-04-19 | | Release date: | 2011-05-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Unexpected binding orientation of bulky-B-ring anti-androgens and implications for future drug targets.
J.Med.Chem., 54, 2011
|
|
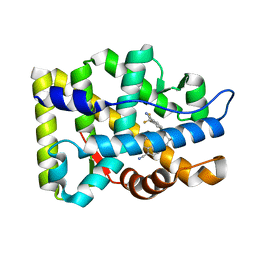
4TUJ
 
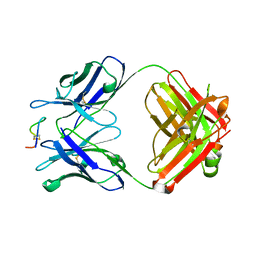 | | Crystal structure of monoclonal antibody against neuroblastoma associated antigen. | | Descriptor: | Heavy chain of monoclonal antibody against neuroblastoma associated antigen, Light chain of monoclonal antibody against neuroblastoma associated antigen, peptide1 | | Authors: | Grudnik, P, Golik, P, Horwacik, I, Zdzalik, M, Rokita, H, Dubin, G. | | Deposit date: | 2014-06-24 | | Release date: | 2015-07-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural Basis of GD2 Ganglioside and Mimetic Peptide Recognition by 14G2a Antibody.
Mol.Cell Proteomics, 14, 2015
|
|
8UPI
 
 | | Structure of a periplasmic peptide binding protein from Mesorhizobium sp. AP09 bound to aminoserine | | Descriptor: | 1,2-ETHANEDIOL, AMINOSERINE, CALCIUM ION, ... | | Authors: | Frkic, R.L, Smith, O.B, Rahman, M, Kaczmarski, J.A, Jackson, C.J. | | Deposit date: | 2023-10-22 | | Release date: | 2023-11-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Identification and Characterization of a Bacterial Periplasmic Solute Binding Protein That Binds l-Amino Acid Amides.
Biochemistry, 63, 2024
|
|
2VDR
 
 | | Integrin AlphaIIbBeta3 Headpiece Bound to a chimeric Fibrinogen Gamma chain peptide, LGGAKQRGDV | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, FIBRINOGEN, ... | | Authors: | Springer, T.A, Zhu, J, Xiao, T. | | Deposit date: | 2007-10-10 | | Release date: | 2008-09-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis for Distinctive Recognition of Fibrinogen Gammac Peptide by the Platelet Integrin Alphaiibbeta3.
J.Cell Biol., 182, 2008
|
|
3QM4
 
 | | Human Cytochrome P450 (CYP) 2D6 - Prinomastat Complex | | Descriptor: | Cytochrome P450 2D6, NICKEL (II) ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Wang, A, Stout, C.D, Johnson, E.F. | | Deposit date: | 2011-02-03 | | Release date: | 2012-02-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal Structure of Human Cytochrome P450 2D6 with Prinomastat Bound.
J.Biol.Chem., 287, 2012
|
|
8B29
 
 | | Human carbonic anhydrase II containing 6-fluorotryptophanes. | | Descriptor: | Carbonic anhydrase 2, ZINC ION | | Authors: | Pham, L.B.T, Costantino, A, Barbieri, L, Calderone, V, Luchinat, E, Banci, L. | | Deposit date: | 2022-09-13 | | Release date: | 2023-01-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Direct Expression of Fluorinated Proteins in Human Cells for 19 F In-Cell NMR Spectroscopy.
J.Am.Chem.Soc., 145, 2023
|
|
1B2J
 
 | | CLOSTRIDIUM PASTEURIANUM RUBREDOXIN G43A MUTANT | | Descriptor: | FE (III) ION, PROTEIN (RUBREDOXIN) | | Authors: | Maher, M.J, Guss, J.M, Wilce, M.C.J, Wedd, A.G. | | Deposit date: | 1998-11-27 | | Release date: | 1999-05-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Rubredoxin from Clostridium pasteurianum. Structures of G10A, G43A and G10VG43A mutant proteins. Mutation of conserved glycine 10 to valine causes the 9-10 peptide link to invert.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
3QO4
 
 | | The Crystal Structure of Death Receptor 6 | | Descriptor: | ACETATE ION, SULFATE ION, Tumor necrosis factor receptor superfamily member 21 | | Authors: | Kuester, M, Kemmerzehl, S, Dahms, S.O, Roeser, D, Than, M.E. | | Deposit date: | 2011-02-09 | | Release date: | 2011-05-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of death receptor 6 (DR6): a potential receptor of the amyloid precursor protein (APP).
J.Mol.Biol., 409, 2011
|
|
1B13
 
 | | CLOSTRIDIUM PASTEURIANUM RUBREDOXIN G10A MUTANT | | Descriptor: | FE (III) ION, PROTEIN (RUBREDOXIN) | | Authors: | Maher, M.J, Guss, J.M, Wilce, M.C.J, Wedd, A.G. | | Deposit date: | 1998-11-26 | | Release date: | 1999-05-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Rubredoxin from Clostridium pasteurianum. Structures of G10A, G43A and G10VG43A mutant proteins. Mutation of conserved glycine 10 to valine causes the 9-10 peptide link to invert.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1B2O
 
 | | CLOSTRIDIUM PASTEURIANUM RUBREDOXIN G10VG43A MUTANT | | Descriptor: | FE (III) ION, PROTEIN (RUBREDOXIN) | | Authors: | Maher, M.J, Guss, J.M, Wilce, M.C.J, Wedd, A.G. | | Deposit date: | 1998-11-30 | | Release date: | 1999-05-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Rubredoxin from Clostridium pasteurianum. Structures of G10A, G43A and G10VG43A mutant proteins. Mutation of conserved glycine 10 to valine causes the 9-10 peptide link to invert.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
4MQ7
 
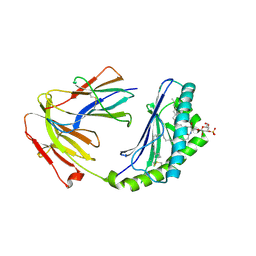 | | Structure of human CD1d-sulfatide | | Descriptor: | (15Z)-N-((1S,2R,3E)-2-HYDROXY-1-{[(3-O-SULFO-BETA-D-GALACTOPYRANOSYL)OXY]METHYL}HEPTADEC-3-ENYL)TETRACOS-15-ENAMIDE, 2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d, ... | | Authors: | Luoma, A.M, Adams, E.J. | | Deposit date: | 2013-09-15 | | Release date: | 2013-12-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6032 Å) | | Cite: | Crystal Structure of V delta 1 T Cell Receptor in Complex with CD1d-Sulfatide Shows MHC-like Recognition of a Self-Lipid by Human gamma delta T Cells.
Immunity, 39, 2013
|
|
4MMX
 
 | | Integrin AlphaVBeta3 ectodomain bound to the tenth domain of Fibronectin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fibronectin, ... | | Authors: | van Agthoven, J, Xiong, J, Arnaout, M.A. | | Deposit date: | 2013-09-09 | | Release date: | 2014-03-26 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (3.32 Å) | | Cite: | Structural basis for pure antagonism of integrin alpha V beta 3 by a high-affinity form of fibronectin.
Nat.Struct.Mol.Biol., 21, 2014
|
|
5UIW
 
 | | Crystal Structure of CC Chemokine Receptor 5 (CCR5) in complex with high potency HIV entry inhibitor 5P7-CCL5 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, C-C chemokine receptor type 5,Rubredoxin chimera, C-C motif chemokine 5, ... | | Authors: | Zheng, Y, Qin, L, Han, G.W, Gustavsson, M, Kawamura, T, Stevens, R.C, Cherezov, V, Kufareva, I, Handel, T.M. | | Deposit date: | 2017-01-15 | | Release date: | 2017-06-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.204 Å) | | Cite: | Structure of CC Chemokine Receptor 5 with a Potent Chemokine Antagonist Reveals Mechanisms of Chemokine Recognition and Molecular Mimicry by HIV.
Immunity, 46, 2017
|
|
3H8K
 
 | | Crystal structure of Ube2g2 complxed with the G2BR domain of gp78 at 1.8-A resolution | | Descriptor: | Autocrine motility factor receptor, isoform 2, Ubiquitin-conjugating enzyme E2 G2 | | Authors: | Kalathur, R.C, Das, R, Li, J, Byrd, R.A, Ji, X. | | Deposit date: | 2009-04-29 | | Release date: | 2009-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Allosteric activation of E2-RING finger-mediated ubiquitylation by a structurally defined specific E2-binding region of gp78.
Mol.Cell, 34, 2009
|
|
1Z4N
 
 | | Structure of beta-phosphoglucomutase with inhibitor bound alpha-galactose 1-phosphate cocrystallized with Fluoride | | Descriptor: | 1-O-phosphono-alpha-D-galactopyranose, Beta-phosphoglucomutase, MAGNESIUM ION | | Authors: | Tremblay, L.W, Zhang, G, Dai, J, Dunaway-Mariano, D, Allen, K.N. | | Deposit date: | 2005-03-16 | | Release date: | 2005-04-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Chemical Confirmation of a Pentavalent Phosphorane in Complex with beta-Phosphoglucomutase
J.Am.Chem.Soc., 127, 2005
|
|
2Q6N
 
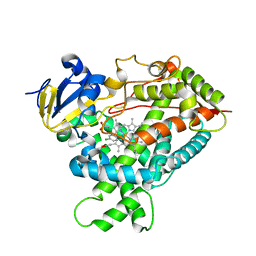 | | Structure of Cytochrome P450 2B4 with Bound 1-(4-cholorophenyl)imidazole | | Descriptor: | 1-(4-CHLOROPHENYL)-1H-IMIDAZOLE, Cytochrome P450 2B4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Zhao, Y, Sun, L, Muralidhara, B.K, Kumar, S, White, M.A, Stout, C.D, Halpert, J.R. | | Deposit date: | 2007-06-05 | | Release date: | 2007-11-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural and thermodynamic consequences of 1-(4-chlorophenyl)imidazole binding to cytochrome P450 2B4.
Biochemistry, 46, 2007
|
|
1BCH
 
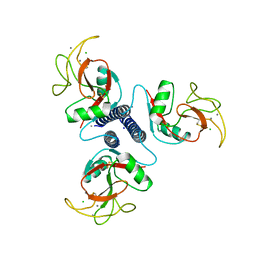 | |
7E2R
 
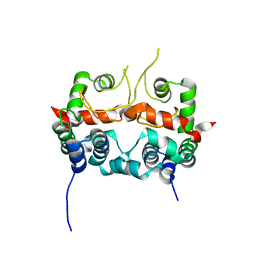 | | The ligand-free structure of Arabidopsis thaliana GUN4 | | Descriptor: | Tetrapyrrole-binding protein, chloroplastic | | Authors: | Liu, L, Hu, J. | | Deposit date: | 2021-02-07 | | Release date: | 2021-08-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of bilin binding by the chlorophyll biosynthesis regulator GUN4.
Protein Sci., 30, 2021
|
|
1BCY
 
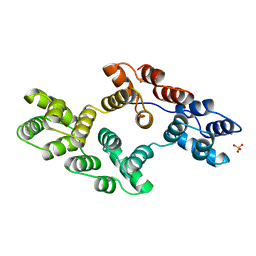 | | RECOMBINANT RAT ANNEXIN V, T72K MUTANT | | Descriptor: | ANNEXIN V, CALCIUM ION, SULFATE ION | | Authors: | Mo, Y.D, Swairjo, M.A, Li, C.W, Head, J.F, Seaton, B.A. | | Deposit date: | 1998-05-04 | | Release date: | 1998-11-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Mutational and crystallographic analyses of interfacial residues in annexin V suggest direct interactions with phospholipid membrane components.
Biochemistry, 37, 1998
|
|
3UN8
 
 | | Yeast 20S proteasome in complex with PR-957 (epoxide) | | Descriptor: | 2-(acetylamino)-4,5-anhydro-1,2-dideoxy-4-methyl-1-phenyl-D-xylitol, Proteasome component C1, Proteasome component C11, ... | | Authors: | Huber, E, Basler, M, Schwab, R, Heinemeyer, W, Kirk, C, Groettrup, M, Groll, M. | | Deposit date: | 2011-11-15 | | Release date: | 2012-02-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Immuno- and constitutive proteasome crystal structures reveal differences in substrate and inhibitor specificity.
Cell(Cambridge,Mass.), 148, 2012
|
|
2VZ3
 
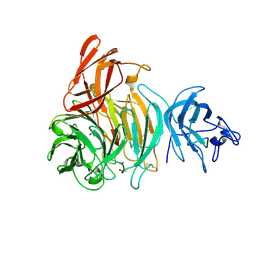 | | bleached galactose oxidase | | Descriptor: | ACETATE ION, COPPER (II) ION, GALACTOSE OXIDASE | | Authors: | Rogers, M.S, Hurtado-Guerrero, R, Firbank, S.J, Halcrow, M.A, Dooley, D.M, Phillips, S.E.V, Knowles, P.F, McPherson, M.J. | | Deposit date: | 2008-07-29 | | Release date: | 2008-09-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cross-Link Formation of the Cysteine 228-Tyrosine 272 Catalytic Cofactor of Galactose Oxidase Does not Require Dioxygen.
Biochemistry, 47, 2008
|
|
4D6Z
 
 | |
4D7D
 
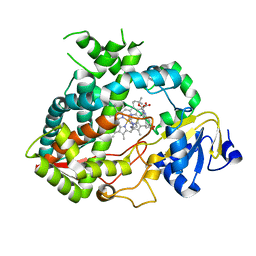 | | Cytochrome P450 3A4 bound to an inhibitor | | Descriptor: | CYTOCHROME P450 3A4, PROTOPORPHYRIN IX CONTAINING FE, tert-butyl [(2S)-1-(1H-indol-3-yl)-3-({3-oxo-3-[(pyridin-3-ylmethyl)amino]propyl}sulfanyl)propan-2-yl]carbamate | | Authors: | Sevrioukova, I, Poulos, T. | | Deposit date: | 2014-11-22 | | Release date: | 2015-09-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Structure-Based Inhibitor Design for Evaluation of a Cyp3A4 Pharmacophore Model.
J.Med.Chem., 59, 2016
|
|
