6TNN
 
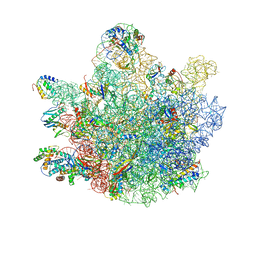 | | Mini-RNase III (Mini-III) bound to 50S ribosome with precursor 23S rRNA | | Descriptor: | 50S ribosomal protein L10, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Oerum, S, Dendooven, T, Gilet, L, Catala, M, Degut, C, Trinquier, A, Barraud, P, Luisi, B, Condon, C, Tisne, C. | | Deposit date: | 2019-12-09 | | Release date: | 2020-09-30 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Structures of B. subtilis Maturation RNases Captured on 50S Ribosome with Pre-rRNAs.
Mol.Cell, 80, 2020
|
|
7FYV
 
 | | Crystal Structure of human FABP4 in complex with 5-(4-chlorophenyl)-9-methyl-3-oxa-4-azatricyclo[5.2.1.02,6]dec-4-ene-8-carboxylic acid, i.e. SMILES [C@H]12[C@H]3[C@@H]([C@@H]([C@@H]([C@H]1ON=C2c1ccc(cc1)Cl)C3)C)C(=O)O with IC50=5.6 microM | | Descriptor: | (3aR,4R,5R,6S,7R,7aR)-3-(4-chlorophenyl)-6-methyl-3a,4,5,6,7,7a-hexahydro-4,7-methano-1,2-benzoxazole-5-carboxylic acid, 1,2-ETHANEDIOL, Fatty acid-binding protein, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Wickens, J, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
5K0J
 
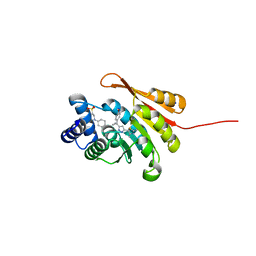 | | Crystal Structure of COMT in complex with 5-[5-[1-(4-methoxyphenyl)cyclopropyl]-1H-pyrazol-3-yl]-2,4-dimethyl-1,3-thiazole | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, 5-{3-[1-(4-methoxyphenyl)cyclopropyl]-1H-pyrazol-5-yl}-2,4-dimethyl-1,3-thiazole, Catechol O-methyltransferase, ... | | Authors: | Ehler, A, Rodriguez-Sarmiento, R.M, Rudolph, M.G. | | Deposit date: | 2016-05-17 | | Release date: | 2016-09-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Design of Potent and Druglike Nonphenolic Inhibitors for Catechol O-Methyltransferase Derived from a Fragment Screening Approach Targeting the S-Adenosyl-l-methionine Pocket.
J. Med. Chem., 59, 2016
|
|
9FT7
 
 | |
8PW7
 
 | | A respirasome from murine liver | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Vercellino, I, Sazanov, L.A. | | Deposit date: | 2023-07-19 | | Release date: | 2024-04-24 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | SCAF1 drives the compositional diversity of mammalian respirasomes.
Nat.Struct.Mol.Biol., 31, 2024
|
|
5I63
 
 | |
7G01
 
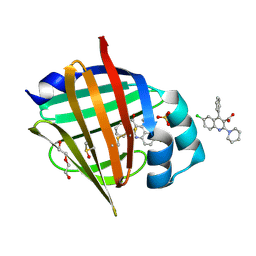 | | Crystal Structure of human FABP5 in complex with 6-chloro-4-phenyl-2-piperidin-1-ylquinoline-3-carboxylic acid, i.e. SMILES n1c(c(c(c2c1ccc(c2)Cl)c1ccccc1)C(=O)O)N1CCCCC1 with IC50=1.1 microM | | Descriptor: | 6-chloranyl-4-phenyl-2-piperidin-1-yl-quinoline-3-carboxylic acid, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Ceccarelli-Simona, M, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
9CAI
 
 | | High-resolution C. elegans 80S ribosome structure - class 1 | | Descriptor: | 18S rRNA, 28S rRNA, 4-{(2R)-2-[(1S,3S,5S)-3,5-dimethyl-2-oxocyclohexyl]-2-hydroxyethyl}piperidine-2,6-dione, ... | | Authors: | Sehgal, E, Serrao, V.H.B, Arribere, J. | | Deposit date: | 2024-06-17 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | High-resolution reconstruction of a C. elegans ribosome sheds light on evolutionary dynamics and tissue specificity.
Rna, 30, 2024
|
|
3JZI
 
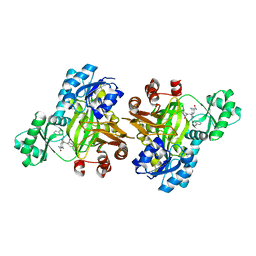 | | Crystal structure of biotin carboxylase from E. Coli in complex with benzimidazole series | | Descriptor: | 7-amino-2-[(2-chlorobenzyl)amino]-1-{[(1S,2S)-2-hydroxycycloheptyl]methyl}-1H-benzimidazole-5-carboxamide, Biotin carboxylase | | Authors: | Cheng, C, Shipps, G.W, Yang, Z, Sun, B, Kawahata, N, Soucy, K, Soriano, A, Orth, P, Xiao, L, Mann, P, Black, T. | | Deposit date: | 2009-09-23 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Discovery and optimization of antibacterial AccC inhibitors.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
4ZKO
 
 | |
7FZO
 
 | | Crystal Structure of human FABP4 in complex with 4-oxo-3-(2-phenylethyl)-10-oxa-3-azatricyclo[5.2.1.01,5]dec-8-ene-6-carboxylic acid, i.e. SMILES [C@H]12[C@@]3(O[C@H]([C@H]2C(=O)O)C=C3)CN(C1=O)CCc1ccccc1 with IC50=3.5 microM | | Descriptor: | (3aR,6S,7R,7aS)-1-oxo-2-(2-phenylethyl)-1,2,3,6,7,7a-hexahydro-3a,6-epoxyisoindole-7-carboxylic acid, FORMIC ACID, Fatty acid-binding protein, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Brunner, M, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
6JAQ
 
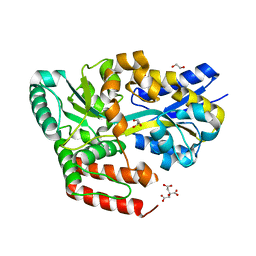 | | Crystal structure of ABC transporter alpha-glycoside-binding mutant protein R356A in complex with glucose | | Descriptor: | 1,2-ETHANEDIOL, ABC transporter, periplasmic substrate-binding protein, ... | | Authors: | Kanaujia, S.P, Chandravanshi, M, Gogoi, P. | | Deposit date: | 2019-01-24 | | Release date: | 2019-10-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and thermodynamic correlation illuminates the selective transport mechanism of disaccharide alpha-glycosides through ABC transporter.
Febs J., 287, 2020
|
|
6ROZ
 
 | |
7G02
 
 | | Crystal Structure of human FABP4 in complex with 7-methoxy-1-(3-methoxyphenyl)naphthalene-2-carboxylic acid, i.e. SMILES c1(c2c(ccc1C(=O)O)ccc(c2)OC)c1cc(ccc1)OC with IC50=0.045 microM | | Descriptor: | (1M)-7-methoxy-1-(3-methoxyphenyl)naphthalene-2-carboxylic acid, 1,2-ETHANEDIOL, FORMIC ACID, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Guthrie-Robert, W, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
6MGR
 
 | | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Campylobacter jejuni in the complex with inhibitor Oxanosine monophosphate | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 5-[(Z)-(aminomethylidene)amino]-1-(5-O-phosphono-beta-D-ribofuranosyl)-1H-imidazole-4-carboxylic acid, CHLORIDE ION, ... | | Authors: | Kim, Y, Maltseva, N, Yu, R, Hedstrom, L, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-09-14 | | Release date: | 2018-10-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Campylobacter jejuni in the complex with inhibitor Oxanosine Monophosphate
To Be Published
|
|
9BJ1
 
 | | Crystal structure of inhibitor GNE-6893 bound to HPK1 | | Descriptor: | (4S,5R,7R,11aP)-10-{[(3R)-3-hydroxy-1-methyl-2-oxopyrrolidin-3-yl]ethynyl}-N~3~-methyl-6,7-dihydro-5H-5,7-methanoimidazo[2,1-a][2]benzazepine-2,3-dicarboxamide, (9S)-2-{[(6P)-8-amino-6-(5-amino-4-methylpyridin-3-yl)-7-fluoroisoquinolin-3-yl]amino}-6-methyl-5,6-dihydro-4H-pyrazolo[1,5-d][1,4]diazepin-7(8H)-one, 1,2-ETHANEDIOL, ... | | Authors: | Kiefer, J.R, Tellis, J.C, Chan, B.K, Wang, W, Wu, P, Choo, E.F, Heffron, T.P, Wei, B, Siu, M. | | Deposit date: | 2024-04-24 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Discovery of GNE-6893, a Potent, Selective, Orally Bioavailable Small Molecule Inhibitor of HPK1.
Acs Med.Chem.Lett., 15, 2024
|
|
7BHR
 
 | | Crystal structure of MAT2a with triazinone fragment 1 bound in the allosteric site | | Descriptor: | 4-(dimethylamino)-6-ethoxy-1~{H}-1,3,5-triazin-2-one, S-ADENOSYLMETHIONINE, S-adenosylmethionine synthase isoform type-2 | | Authors: | Schimpl, M, De Fusco, C, Borjesson, U, Cheung, T, Collie, I, Evans, L, Narasimhan, P, Stubbs, C, Vazquez-Chantada, M, Wagner, D.J, Grondine, M, Tentarelli, S, Underwood, E, Argyrou, A, Bagal, S, Chiarparin, E, Robb, G, Scott, J.S. | | Deposit date: | 2021-01-11 | | Release date: | 2021-04-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Fragment-Based Design of a Potent MAT2a Inhibitor and in Vivo Evaluation in an MTAP Null Xenograft Model.
J.Med.Chem., 64, 2021
|
|
7FYX
 
 | | Crystal Structure of human FABP4 in complex with 2-[(3-phenylthiophen-2-yl)carbamoyl]cyclopentene-1-carboxylic acid, i.e. SMILES S1C(=C(C=C1)c1ccccc1)NC(=O)C1=C(CCC1)C(=O)O with IC50=0.201 microM | | Descriptor: | 2-[(3-phenylthiophen-2-yl)carbamoyl]cyclopent-1-ene-1-carboxylic acid, Fatty acid-binding protein, adipocyte, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Ceccarelli-Simona, M, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
8PW6
 
 | | C respirasome from murine liver | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Vercellino, I, Sazanov, L.A. | | Deposit date: | 2023-07-19 | | Release date: | 2024-04-24 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | SCAF1 drives the compositional diversity of mammalian respirasomes.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8PW5
 
 | | CS respirasome from murine liver | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Vercellino, I, Sazanov, L.A. | | Deposit date: | 2023-07-19 | | Release date: | 2024-04-24 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | SCAF1 drives the compositional diversity of mammalian respirasomes.
Nat.Struct.Mol.Biol., 31, 2024
|
|
9C4R
 
 | | Crystal structure of wild-type arabidopsis thaliana acetohydroxyacid synthase in complex with newly designed herbicide CMO | | Descriptor: | 2-(2-chloroethoxy)-N-[(4-methoxy-6-methylpyrimidin-2-yl)carbamoyl]benzene-1-sulfonamide, 2-[3-[(4-azanyl-2-methyl-pyrimidin-5-yl)methyl]-2-[(1~{S})-1-(dioxidanyl)-1-oxidanyl-ethyl]-4-methyl-1,3-thiazol-5-yl]ethyl phosphono hydrogen phosphate, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, ... | | Authors: | Cheng, Y, Guddat, L.W. | | Deposit date: | 2024-06-05 | | Release date: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Crystal Structures of Arabidopsis thaliana Acetohydroxyacid Synthase in Complex with the Herbicide Triasulfuron and Two Analogues with Herbicidal Activity in Field Trials.
J.Agric.Food Chem., 2024
|
|
3K66
 
 | | X-ray crystal structure of the E2 domain of C. elegans APL-1 | | Descriptor: | Beta-amyloid-like protein | | Authors: | Hoopes, J.T, Ha, Y. | | Deposit date: | 2009-10-08 | | Release date: | 2009-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural characterization of the E2 domain of APL-1, a Caenorhabditis elegans homolog of human amyloid precursor protein, and its heparin binding site
J.Biol.Chem., 285, 2010
|
|
1O4F
 
 | | CRYSTAL STRUCTURE OF SH2 IN COMPLEX WITH RU79073. | | Descriptor: | 1,2,3,4-TETRAHYDROQUINOLIN-8-YL DIHYDROGEN PHOSPHATE, PROTO-ONCOGENE TYROSINE-PROTEIN KINASE SRC | | Authors: | Lange, G, Loenze, P, Liesum, A. | | Deposit date: | 2003-06-15 | | Release date: | 2004-02-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Requirements for specific binding of low affinity inhibitor fragments to the SH2 domain of (pp60)Src are identical to those for high affinity binding of full length inhibitors.
J.Med.Chem., 46, 2003
|
|
4J8P
 
 | |
9C4Q
 
 | | Crystal structure of wild-type arabidopsis thaliana acetohydroxyacid synthase in complex with newly designed herbicide FMO | | Descriptor: | 2-(2-fluoroethoxy)-N-[(4-methoxy-6-methylpyrimidin-2-yl)carbamoyl]benzene-1-sulfonamide, 2-[3-[(4-azanyl-2-methyl-pyrimidin-5-yl)methyl]-2-[(1~{S})-1-(dioxidanyl)-1-oxidanyl-ethyl]-4-methyl-1,3-thiazol-5-yl]ethyl phosphono hydrogen phosphate, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, ... | | Authors: | Cheng, Y, Guddat, L.W. | | Deposit date: | 2024-06-05 | | Release date: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Crystal Structures of Arabidopsis thaliana Acetohydroxyacid Synthase in Complex with the Herbicide Triasulfuron and Two Analogues with Herbicidal Activity in Field Trials.
J.Agric.Food Chem., 2024
|
|
