1LHE
 
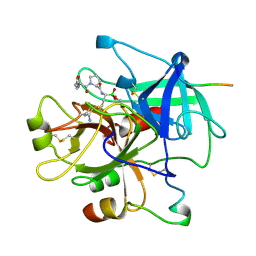 | | HUMAN ALPHA-THROMBIN COMPLEXED WITH AC-(D)PHE-PRO-BORO-N-BUTYL-AMIDINO-GLYCINE-OH | | Descriptor: | AC-(D)PHE-PRO-BORO-N-BUTYL-AMIDINO-GLYCINE-OH, ALPHA-THROMBIN, HIRUDIN | | Authors: | Weber, P.C, Lee, S.L, Lewandowski, F.A, Schadt, M.C, Chang, C.H, Kettner, C.A. | | Deposit date: | 1994-12-27 | | Release date: | 1996-11-08 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Kinetic and crystallographic studies of thrombin with Ac-(D)Phe-Pro-boroArg-OH and its lysine, amidine, homolysine, and ornithine analogs.
Biochemistry, 34, 1995
|
|
1DTX
 
 | |
1LHG
 
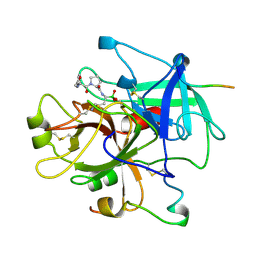 | | HUMAN ALPHA-THROMBIN COMPLEXED WITH AC-(D)PHE-PRO-BOROORNITHINE-OH | | Descriptor: | AC-(D)PHE-PRO-BOROHOMOORNITHINE-OH, ALPHA-THROMBIN, HIRUDIN | | Authors: | Weber, P.C, Lee, S.L, Lewandowski, F.A, Schadt, M.C, Chang, C.H, Kettner, C.A. | | Deposit date: | 1994-12-27 | | Release date: | 1996-11-08 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Kinetic and crystallographic studies of thrombin with Ac-(D)Phe-Pro-boroArg-OH and its lysine, amidine, homolysine, and ornithine analogs.
Biochemistry, 34, 1995
|
|
1IY5
 
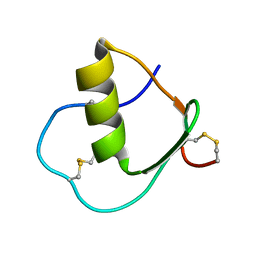 | | Solution structure of wild type OMSVP3 | | Descriptor: | OMSVP3 | | Authors: | Hemmi, H, Kumazaki, T, Yamazaki, T, Kojima, S, Yoshida, T, Kyogoku, Y, Katsu, M, Yokosawa, H, Miura, K, Kobayashi, Y. | | Deposit date: | 2002-07-23 | | Release date: | 2003-03-11 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Inhibitory Specificity Change of Ovomucoid Third Domain of the Silver Pheasant upon Introduction of an Engineered Cys14-Cys39 Bond
BIOCHEMISTRY, 42, 2003
|
|
1LMW
 
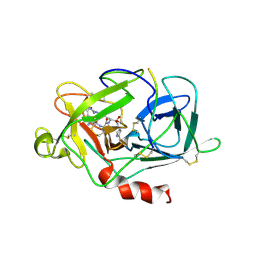 | | LMW U-PA Structure complexed with EGRCMK (GLU-GLY-ARG Chloromethyl Ketone) | | Descriptor: | L-alpha-glutamyl-N-{(1S)-4-{[amino(iminio)methyl]amino}-1-[(1S)-2-chloro-1-hydroxyethyl]butyl}glycinamide, UROKINASE-TYPE PLASMINOGEN ACTIVATOR | | Authors: | Spraggon, G.S, Phillips, C, Nowak, U.K, Ponting, C.P, Saunders, D, Dobson, C.M, Stuart, D.I, Jones, E.Y. | | Deposit date: | 1995-07-26 | | Release date: | 1996-01-29 | | Last modified: | 2013-02-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of the catalytic domain of human urokinase-type plasminogen activator.
Structure, 3, 1995
|
|
1DXW
 
 | | structure of hetero complex of non structural protein (NS) of hepatitis C virus (HCV) and synthetic peptidic compound | | Descriptor: | N-(tert-butoxycarbonyl)-L-alpha-glutamyl-N-[(1R)-1-(carboxycarbonyl)-3,3-difluoropropyl]-L-leucinamide, SERINE PROTEASE, ZINC ION | | Authors: | Barbato, G, Cicero, D.O, Cordier, F, Narjes, F, Gerlach, B, Sambucini, S, Grzesiek, S, Matassa, V.G, Defrancesco, R, Bazzo, R. | | Deposit date: | 2000-01-17 | | Release date: | 2001-01-12 | | Last modified: | 2020-01-15 | | Method: | SOLUTION NMR | | Cite: | Inhibitor Binding Induces Active Site Stabilisation of the Hcv Ns3 Protein Serine Protease Domain
Embo J., 19, 2000
|
|
1CO7
 
 | |
1DY9
 
 | | Inhibition of the Hepatitis C Virus NS3/4A Protease. The Crystal Structures of Two Protease-Inhibitor Complexes (inhibitor I) | | Descriptor: | N-(tert-butoxycarbonyl)-L-alpha-glutamyl-N-[(1R)-1-(carboxycarbonyl)-3,3-difluoropropyl]-L-leucinamide, NONSTRUCTURAL PROTEIN NS4A (P4), PROTEASE/HELICASE NS3 (P70), ... | | Authors: | Di Marco, S, Rizzi, M, Volpari, C, Walsh, M, Narjes, F, Colarusso, S, De Francesco, R, Matassa, V.G, Sollazzo, M. | | Deposit date: | 2000-01-31 | | Release date: | 2001-01-28 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Inhibition of the Hepatitis C Virus Ns3/4A Protease the Crystal Structures of Two Protease-Inhibitor Complexes
J.Biol.Chem., 275, 2000
|
|
1JPG
 
 | | Crystal Structure Of The LCMV Peptidic Epitope Np396 In Complex With The Murine Class I Mhc Molecule H-2Db | | Descriptor: | BETA-2-MICROGLOBULIN, H-2 CLASS I HISTOCOMPATIBILITY ANTIGEN, D-B ALPHA CHAIN, ... | | Authors: | Ciatto, C, Tissot, A.C, Tschopp, M, Capitani, G, Pecorari, F, Pluckthun, A, Grutter, M.G. | | Deposit date: | 2001-08-02 | | Release date: | 2001-10-24 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Zooming in on the hydrophobic ridge of H-2D(b): implications for the conformational variability of bound peptides.
J.Mol.Biol., 312, 2001
|
|
1DY8
 
 | | Inhibition of the Hepatitis C Virus NS3/4A Protease. The Crystal Structures of Two Protease-Inhibitor Complexes (inhibitor II) | | Descriptor: | N-[(benzyloxy)carbonyl]-L-isoleucyl-N-[(1R)-1-(carboxycarbonyl)-3,3-difluoropropyl]-L-leucinamide, NONSTRUCTURAL PROTEIN NS4A (P4), PROTEASE/HELICASE NS3 (P70) | | Authors: | Di Marco, S, Rizzi, M, Volpari, C, Walsh, M, Narjes, F, Colarusso, S, De Francesco, R, Matassa, V.G, Sollazzo, M. | | Deposit date: | 2000-01-31 | | Release date: | 2001-01-28 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Inhibition of the Hepatitis C Virus Ns3/4A Protease the Crystal Structures of Two Protease-Inhibitor Complexes
J.Biol.Chem., 275, 2000
|
|
1DXP
 
 | | Inhibition of the Hepatitis C Virus NS3/4A Protease. The Crystal Structures of Two Protease-Inhibitor Complexes (apo structure) | | Descriptor: | GLYCEROL, NONSTRUCTURAL PROTEIN NS4A (P4), PROTEASE/HELICASE NS3 (P70), ... | | Authors: | Di Marco, S, Rizzi, M, Volpari, C, Walsh, M, Narjes, F, Colarusso, S, De Francesco, R, Matassa, V.G, Sollazzo, M. | | Deposit date: | 2000-01-13 | | Release date: | 2001-01-12 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Inhibition of the Hepatitis C Virus Ns3/4A Protease the Crystal Structures of Two Protease-Inhibitor Complexes
J.Biol.Chem., 275, 2000
|
|
1ESE
 
 | | THE MOLECULAR MECHANISM OF ENANTIORECOGNITION BY ESTERASES | | Descriptor: | DIETHYL PHOSPHONATE, ESTERASE | | Authors: | Wei, Y, Schottel, J.L, Derewenda, U, Swenson, L, Patkar, S, Derewenda, Z.S. | | Deposit date: | 1994-10-07 | | Release date: | 1995-10-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A novel variant of the catalytic triad in the Streptomyces scabies esterase.
Nat.Struct.Biol., 2, 1995
|
|
3D9T
 
 | |
2SGP
 
 | | PRO 18 VARIANT OF TURKEY OVOMUCOID INHIBITOR THIRD DOMAIN COMPLEXED WITH STREPTOMYCES GRISEUS PROTEINASE B AT PH 6.5 | | Descriptor: | OVOMUCOID INHIBITOR, PHOSPHATE ION, PROTEINASE B | | Authors: | Huang, K, Lu, W, Anderson, S, Laskowski Jr, M, James, M.N.G. | | Deposit date: | 1999-03-25 | | Release date: | 2001-01-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of peptide bonds to inhibitor-protease binding: crystal structures of the turkey ovomucoid third domain backbone variants OMTKY3-Pro18I and OMTKY3-psi[COO]-Leu18I in complex with Streptomyces griseus proteinase B (SGPB) and the structure of the free inhibitor, OMTKY-3-psi[CH2NH2+]-Asp19I
J.Mol.Biol., 305, 2001
|
|
3DA7
 
 | | A conformationally strained, circular permutant of barnase | | Descriptor: | Barnase circular permutant, Barstar | | Authors: | Mitrousis, G, Butler, J, Loh, S.N, Cingolani, G. | | Deposit date: | 2008-05-28 | | Release date: | 2009-04-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural and thermodynamic analysis of a conformationally strained circular permutant of barnase.
Biochemistry, 48, 2009
|
|
3DEJ
 
 | | Crystal Structures of Caspase-3 with Bound Isoquinoline-1,3,4-trione Derivative Inhibitors | | Descriptor: | (1S)-1-(3-chlorophenyl)-2-oxo-2-[(1,3,4-trioxo-1,2,3,4-tetrahydroisoquinolin-5-yl)amino]ethyl acetate, Caspase-3 | | Authors: | Wu, J, Du, J, Li, J, Ding, J. | | Deposit date: | 2008-06-10 | | Release date: | 2008-09-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Isoquinoline-1,3,4-trione Derivatives Inactivate Caspase-3 by Generation of Reactive Oxygen Species
J.Biol.Chem., 283, 2008
|
|
3DNG
 
 | | Crystal structure of the complex between MMP-8 and a non-zinc chelating inhibitor | | Descriptor: | (5S)-5-(2-amino-2-oxoethyl)-4-oxo-N-[(3-oxo-3,4-dihydro-2H-1,4-benzoxazin-6-yl)methyl]-3,4,5,6,7,8-hexahydro[1]benzothieno[2,3-d]pyrimidine-2-carboxamide, CALCIUM ION, Neutrophil collagenase, ... | | Authors: | Pochetti, G, Montanari, R, Mazza, F. | | Deposit date: | 2008-07-02 | | Release date: | 2009-03-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Extra Binding Region Induced by Non-Zinc Chelating Inhibitors into the S(1)' Subsite of Matrix Metalloproteinase 8 (MMP-8)
J.Med.Chem., 52, 2009
|
|
3DA9
 
 | |
3DEH
 
 | | Crystal Structures of Caspase-3 with Bound Isoquinoline-1,3,4-trione Derivative Inhibitors | | Descriptor: | Caspase-3, isoquinoline-1,3,4(2H)-trione | | Authors: | Wu, J, Du, J, Li, J, Ding, J. | | Deposit date: | 2008-06-10 | | Release date: | 2008-09-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Isoquinoline-1,3,4-trione Derivatives Inactivate Caspase-3 by Generation of Reactive Oxygen Species
J.Biol.Chem., 283, 2008
|
|
2RDL
 
 | | Hamster Chymase 2 | | Descriptor: | Chymase 2, METHOXYSUCCINYL-ALA-ALA-PRO-ALA-CHLOROMETHYLKETONE INHIBITOR, SULFATE ION | | Authors: | Spurlino, J, Abad, M, Kervinen, J. | | Deposit date: | 2007-09-24 | | Release date: | 2007-10-30 | | Last modified: | 2012-12-12 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for elastolytic substrate specificity in rodent alpha-chymases.
J.Biol.Chem., 283, 2008
|
|
2RJP
 
 | | Crystal structure of ADAMTS4 with inhibitor bound | | Descriptor: | ADAMTS-4, CALCIUM ION, N-({4'-[(4-isobutyrylphenoxy)methyl]biphenyl-4-yl}sulfonyl)-D-valine, ... | | Authors: | Mosyak, L, Stahl, M, Somers, W. | | Deposit date: | 2007-10-15 | | Release date: | 2007-12-11 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of the two major aggrecan degrading enzymes, ADAMTS4 and ADAMTS5.
Protein Sci., 17, 2008
|
|
6JGJ
 
 | | Crystal structure of the F99S/M153T/V163A/E222Q variant of GFP at 0.78 A | | Descriptor: | Green fluorescent protein, MAGNESIUM ION | | Authors: | Takaba, K, Tai, Y, Hanazono, Y, Miki, K, Takeda, K. | | Deposit date: | 2019-02-14 | | Release date: | 2019-04-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (0.78 Å) | | Cite: | Subatomic resolution X-ray structures of green fluorescent protein.
Iucrj, 6, 2019
|
|
6JGH
 
 | | Crystal structure of the F99S/M153T/V163A/T203I variant of GFP at 0.94 A | | Descriptor: | CHLORIDE ION, Green fluorescent protein | | Authors: | Eki, H, Tai, Y, Takaba, K, Hanazono, Y, Miki, K, Takeda, K. | | Deposit date: | 2019-02-14 | | Release date: | 2019-04-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (0.94 Å) | | Cite: | Subatomic resolution X-ray structures of green fluorescent protein.
Iucrj, 6, 2019
|
|
3QVI
 
 | | Crystal structure of KNI-10395 bound histo-aspartic protease (HAP) from Plasmodium falciparum | | Descriptor: | (4R)-N-[(1S,2R)-2-hydroxy-2,3-dihydro-1H-inden-1-yl]-3-[(2S,3S)-2-hydroxy-3-{[S-methyl-N-(phenylacetyl)-L-cysteinyl]amino}-4-phenylbutanoyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxamide, 1,2-ETHANEDIOL, 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, ... | | Authors: | Bhaumik, P, Gustchina, A, Wlodawer, A. | | Deposit date: | 2011-02-25 | | Release date: | 2011-10-12 | | Last modified: | 2013-09-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into the activation and inhibition of histo-aspartic protease from Plasmodium falciparum.
Biochemistry, 50, 2011
|
|
5FYN
 
 | | Sub-tomogram averaging of Tula virus glycoprotein spike | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PUUMALA VIRUS GN GLYCOPROTEIN, alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, S, Rissanen, I, Zeltina, A, Hepojoki, J, Raghwani, J, Harlos, K, Pybus, O.G, Huiskonen, J.T, Bowden, T.A. | | Deposit date: | 2016-03-08 | | Release date: | 2016-06-08 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (15.6 Å) | | Cite: | A Molecular-Level Account of the Antigenic Hantaviral Surface.
Cell Rep., 15, 2016
|
|
