4QPL
 
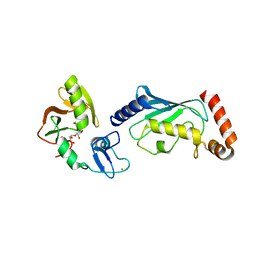 | | Crystal structure of RNF146(RING-WWE)/UbcH5a/iso-ADPr complex | | Descriptor: | 2'-O-(5-O-phosphono-alpha-D-ribofuranosyl)adenosine 5'-(dihydrogen phosphate), E3 ubiquitin-protein ligase RNF146, Ubiquitin-conjugating enzyme E2 D1, ... | | Authors: | Wang, Z, DaRosa, P.A, Klevit, R.E, Xu, W. | | Deposit date: | 2014-06-23 | | Release date: | 2014-10-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Allosteric activation of the RNF146 ubiquitin ligase by a poly(ADP-ribosyl)ation signal.
Nature, 517, 2015
|
|
8V1W
 
 | |
8V1V
 
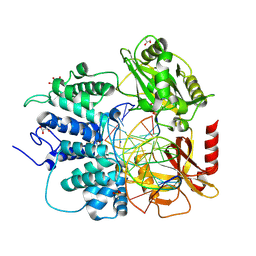 | |
8V1U
 
 | |
4KCD
 
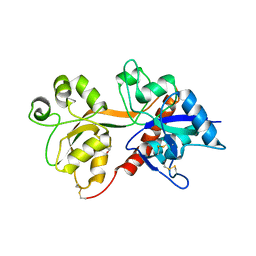 | | Crystal Structure of the NMDA Receptor GluN3A Ligand Binding Domain Apo State | | Descriptor: | GLYCEROL, Glutamate receptor ionotropic, NMDA 3A | | Authors: | Yao, Y, Lau, A.Y, Mayer, M.L. | | Deposit date: | 2013-04-24 | | Release date: | 2013-07-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Conformational Analysis of NMDA Receptor GluN1, GluN2, and GluN3 Ligand-Binding Domains Reveals Subtype-Specific Characteristics.
Structure, 21, 2013
|
|
6RCY
 
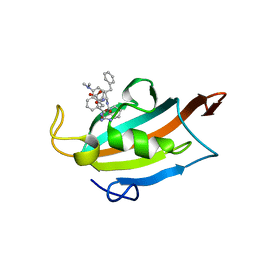 | |
5O6T
 
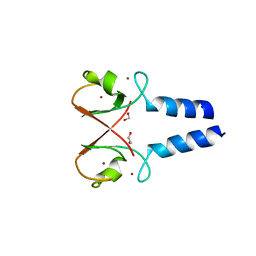 | | BIRC4 RING in complex with dimeric ubiquitin variant | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase XIAP, Polyubiquitin-B, ... | | Authors: | Gabrielsen, M, Buetow, L, Huang, D.T. | | Deposit date: | 2017-06-07 | | Release date: | 2017-11-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | A General Strategy for Discovery of Inhibitors and Activators of RING and U-box E3 Ligases with Ubiquitin Variants.
Mol. Cell, 68, 2017
|
|
7DPW
 
 | | Structural basis for ligand binding modes of CTP synthase | | Descriptor: | CTP synthase, CYTIDINE-5'-TRIPHOSPHATE, MAGNESIUM ION | | Authors: | Liu, J.L, Zhou, X, Guo, C.J, Chang, C.C. | | Deposit date: | 2020-12-21 | | Release date: | 2021-09-15 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | Structural basis for ligand binding modes of CTP synthase.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
4KCC
 
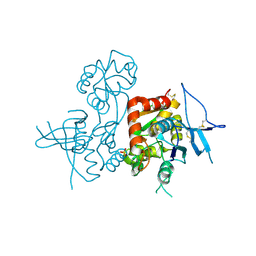 | | Crystal Structure of the NMDA Receptor GluN1 Ligand Binding Domain Apo State | | Descriptor: | Glutamate receptor ionotropic, NMDA 1, PHOSPHATE ION | | Authors: | Berger, A.J, Lau, A.Y, Mayer, M.L. | | Deposit date: | 2013-04-24 | | Release date: | 2013-07-31 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.894 Å) | | Cite: | Conformational Analysis of NMDA Receptor GluN1, GluN2, and GluN3 Ligand-Binding Domains Reveals Subtype-Specific Characteristics.
Structure, 21, 2013
|
|
2I1P
 
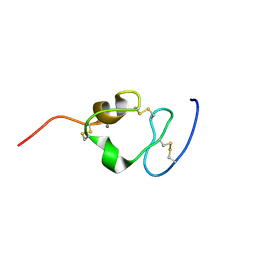 | | Solution structure of the twelfth cysteine-rich ligand-binding repeat in rat megalin | | Descriptor: | CALCIUM ION, Low-density lipoprotein receptor-related protein 2 | | Authors: | Wolf, C.A, Dancea, F, Shi, M, Bade-Noskova, V, Rueterjans, H, Kerjaschki, D, Luecke, C. | | Deposit date: | 2006-08-14 | | Release date: | 2007-02-13 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the twelfth cysteine-rich ligand-binding repeat in rat megalin.
J.Biomol.Nmr, 37, 2007
|
|
8K9I
 
 | | Structure of CUL3-RBX1-KLHL22 complex without CUL3 NA motif | | Descriptor: | Cullin-3, E3 ubiquitin-protein ligase RBX1, N-terminally processed, ... | | Authors: | Wang, W, Ling, L, Dai, Z, Zuo, P, Yin, Y. | | Deposit date: | 2023-08-01 | | Release date: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | A conserved N-terminal motif of CUL3 contributes to assembly and E3 ligase activity of CRL3 KLHL22.
Nat Commun, 15, 2024
|
|
1UOM
 
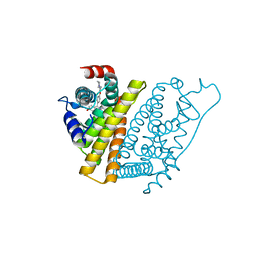 | | The Structure of Estrogen Receptor in Complex with a Selective and Potent Tetrahydroisochiolin Ligand. | | Descriptor: | 2-PHENYL-1-[4-(2-PIPERIDIN-1-YL-ETHOXY)-PHENYL]-1,2,3,4-TETRAHYDRO-ISOQUINOLIN-6-OL, ESTROGEN RECEPTOR | | Authors: | Stark, W, Bischoff, S.F, Buhl, T, Fournier, B, Halleux, C, Kallen, J, Keller, H, Renaud, J. | | Deposit date: | 2003-04-11 | | Release date: | 2003-07-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Estrogen Receptor Modulators: Identification and Structure-Activity Relationships of Potent Eralpha-Selective Tetrahydroisoquinoline Ligands
J.Med.Chem., 46, 2003
|
|
6UML
 
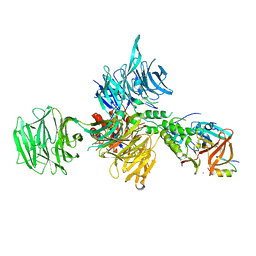 | | Structural Basis for Thalidomide Teratogenicity Revealed by the Cereblon-DDB1-SALL4-Pomalidomide Complex | | Descriptor: | DNA damage-binding protein 1, Protein cereblon, S-Pomalidomide, ... | | Authors: | Clayton, T.L, Matyskiela, M.E, Pagarigan, B.E, Tran, E.T, Chamberlain, P.P. | | Deposit date: | 2019-10-09 | | Release date: | 2020-04-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.58 Å) | | Cite: | Crystal structure of the SALL4-pomalidomide-cereblon-DDB1 complex.
Nat.Struct.Mol.Biol., 27, 2020
|
|
2P15
 
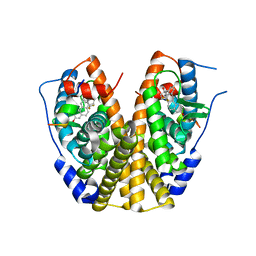 | | Crystal structure of the ER alpha ligand binding domain with the agonist ortho-trifluoromethylphenylvinyl estradiol | | Descriptor: | (17BETA)-17-{(E)-2-[2-(TRIFLUOROMETHYL)PHENYL]VINYL}ESTRA-1(10),2,4-TRIENE-3,17-DIOL, Estrogen receptor, GRIP peptide | | Authors: | Bruning, J.B, Nettles, K.W, Greene, G.L, Kim, Y. | | Deposit date: | 2007-03-02 | | Release date: | 2007-05-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural plasticity in the oestrogen receptor ligand-binding domain.
Embo Rep., 8, 2007
|
|
3VRU
 
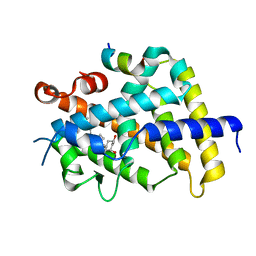 | | VDR ligand binding domain in complex with 2-Methylidene-19,24-dinor-1alpha,25-dihydroxy vitaminD3 | | Descriptor: | (1R,3R,7E,17beta)-17-[(2R)-5-hydroxy-5-methylhexan-2-yl]-2-methylidene-9,10-secoestra-5,7-diene-1,3-diol, 13-meric peptide from Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | Authors: | Yoshimoto, N, Inaba, Y, Itoh, T, Nakabayashi, M, Ito, N, Yamamoto, K. | | Deposit date: | 2012-04-14 | | Release date: | 2012-05-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Butyl pocket formation in the vitamin d receptor strongly affects the agonistic or antagonistic behavior of ligands
J.Med.Chem., 55, 2012
|
|
1QKU
 
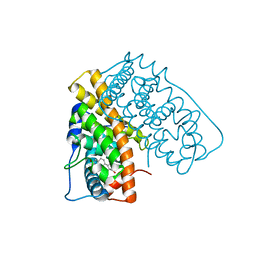 | | WILD TYPE ESTROGEN NUCLEAR RECEPTOR LIGAND BINDING DOMAIN COMPLEXED WITH ESTRADIOL | | Descriptor: | ESTRADIOL, ESTRADIOL RECEPTOR | | Authors: | Ruff, M, Gangloff, M, Eiler, S, Duclaud, S, Wurtz, J.M, Moras, D. | | Deposit date: | 1999-08-05 | | Release date: | 2000-08-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of a mutant hERalpha ligand-binding domain reveals key structural features for the mechanism of partial agonism.
J. Biol. Chem., 276, 2001
|
|
7NPC
 
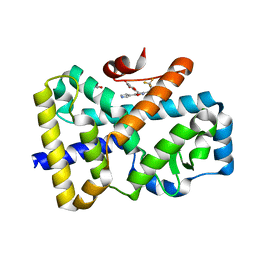 | | ROR(gamma)t ligand binding domain in complex with allosteric ligand FM156 | | Descriptor: | 4-[[3-[2-chloranyl-6-(trifluoromethyl)phenyl]-5-(1~{H}-pyrrol-3-yl)-1,2-oxazol-4-yl]methoxy]benzoic acid, GLYCEROL, Nuclear receptor ROR-gamma | | Authors: | de Vries, R.M.J.M, Meijer, F.A, Brunsveld, L. | | Deposit date: | 2021-02-26 | | Release date: | 2021-06-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structure-Activity Relationship Studies of Trisubstituted Isoxazoles as Selective Allosteric Ligands for the Retinoic-Acid-Receptor-Related Orphan Receptor gamma t.
J.Med.Chem., 64, 2021
|
|
7NP5
 
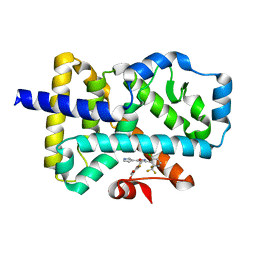 | | ROR(gamma)t ligand binding domain in complex with allosteric ligand FM216 | | Descriptor: | 4-[[3-[2-chloranyl-6-(trifluoromethyl)phenyl]-5-(1~{H}-pyrrol-3-yl)-1,2-oxazol-4-yl]methoxy]-2-fluoranyl-benzoic acid, Nuclear receptor ROR-gamma | | Authors: | Oerlemans, G.J.M, Somsen, B.A, de Vries, R.M.J.M, Meijer, F.A, Brunsveld, L. | | Deposit date: | 2021-02-26 | | Release date: | 2021-06-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure-Activity Relationship Studies of Trisubstituted Isoxazoles as Selective Allosteric Ligands for the Retinoic-Acid-Receptor-Related Orphan Receptor gamma t.
J.Med.Chem., 64, 2021
|
|
7NP6
 
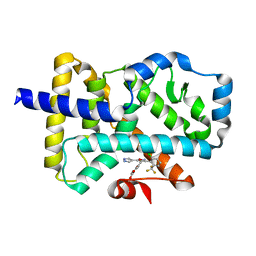 | | ROR(gamma)t ligand binding domain in complex with allosteric ligand FM257 | | Descriptor: | 4-[[3-[2-chloranyl-6-(trifluoromethyl)phenyl]-5-(1~{H}-pyrazol-4-yl)-1,2-oxazol-4-yl]methoxy]benzoic acid, Nuclear receptor ROR-gamma | | Authors: | Oerlemans, G.J.M, Somsen, B.A, de Vries, R.M.J.M, Meijer, F.A, Brunsveld, L. | | Deposit date: | 2021-02-26 | | Release date: | 2021-06-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structure-Activity Relationship Studies of Trisubstituted Isoxazoles as Selective Allosteric Ligands for the Retinoic-Acid-Receptor-Related Orphan Receptor gamma t.
J.Med.Chem., 64, 2021
|
|
2WRZ
 
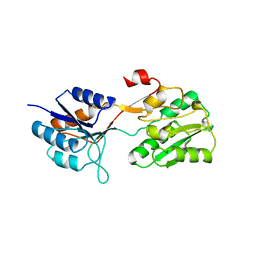 | | Crystal structure of an arabinose binding protein with designed serotonin binding site in open, ligand-free state | | Descriptor: | L-ARABINOSE-BINDING PERIPLASMIC PROTEIN | | Authors: | Schreier, B, Stumpp, C, Wiesner, S, Hocker, B. | | Deposit date: | 2009-09-03 | | Release date: | 2009-10-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Computational Design of Ligand Binding is not a Solved Problem
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2A3Y
 
 | | Pentameric crystal structure of human serum amyloid P-component bound to Bis-1,2-{[(Z)-2carboxy-2-methyl-1,3-dioxane]-5-yloxycarbamoyl}-ethane. | | Descriptor: | BIS-1,2-{[(Z)-2-CARBOXY-2-METHYL-1,3-DIOXANE]-5-YLOXYCARBAMOYL}-ETHANE, CALCIUM ION, Serum amyloid P-component | | Authors: | Ho, J.G, Kitov, P.I, Paszkiewicz, E, Sadowska, J, Bundle, D.R, Ng, K.K. | | Deposit date: | 2005-06-27 | | Release date: | 2005-07-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ligand-assisted Aggregation of Proteins: DIMERIZATION OF SERUM AMYLOID P COMPONENT BY BIVALENT LIGANDS.
J.Biol.Chem., 280, 2005
|
|
3VRW
 
 | | VDR ligand binding domain in complex with 22S-Butyl-2-methylidene-26,27-dimethyl-19,24-dinor-1alpha,25-dihydroxyvitamin D3 | | Descriptor: | (1R,3R,7E,17beta)-17-[(2R,3S)-3-butyl-5-ethyl-5-hydroxyheptan-2-yl]-2-methylidene-9,10-secoestra-5,7-diene-1,3-diol, 13-meric peptide from Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | Authors: | Yoshimoto, N, Inaba, Y, Itoh, T, Nakabayashi, M, Ito, N, Yamamoto, K. | | Deposit date: | 2012-04-16 | | Release date: | 2012-05-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Butyl pocket formation in the vitamin d receptor strongly affects the agonistic or antagonistic behavior of ligands
J.Med.Chem., 55, 2012
|
|
3VRV
 
 | | VDR ligand binding domain in complex with 2-Methylidene-26,27-dimethyl-19,24-dinor-1alpha,25-dihydroxyvitamin D3 | | Descriptor: | (1R,3R,7E,17beta)-17-[(2R)-5-ethyl-5-hydroxyheptan-2-yl]-2-methylidene-9,10-secoestra-5,7-diene-1,3-diol, 13-meric peptide from Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | Authors: | Yoshimoto, N, Inaba, Y, Itoh, T, Nakabayashi, M, Ito, N, Yamamoto, K. | | Deposit date: | 2012-04-14 | | Release date: | 2012-05-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Butyl pocket formation in the vitamin d receptor strongly affects the agonistic or antagonistic behavior of ligands
J.Med.Chem., 55, 2012
|
|
3C7I
 
 | |
3EPE
 
 | |
