6TTF
 
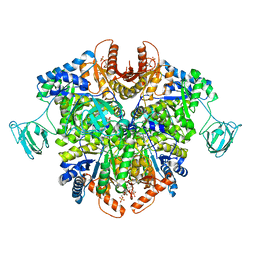 | | PKM2 in complex with Compound 5 | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 5-hydroxynaphthalene-1-sulfonamide, Pyruvate kinase PKM | | Authors: | Saur, M, Hartshorn, M.J, Dong, J, Reeks, J, Bunkoczi, G, Jhoti, H, Williams, P.A. | | Deposit date: | 2019-12-27 | | Release date: | 2020-01-15 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Fragment-based drug discovery using cryo-EM.
Drug Discov Today, 25, 2020
|
|
2P8P
 
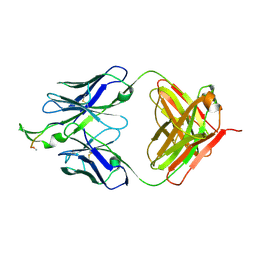 | | Crystal structure of the HIV-1 Cross Neutralizing Monoclonal Antibody 2F5 in complex with gp41 Peptide LELDKWASLW[N-Ac] | | Descriptor: | gp41 peptide, nmAb 2F5, heavy chain, ... | | Authors: | Bryson, S, Julien, J.-P, Pai, E.F. | | Deposit date: | 2007-03-22 | | Release date: | 2007-05-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural details of HIV-1 recognition by the broadly neutralizing monoclonal antibody 2F5: epitope conformation, antigen-recognition loop mobility, and anion-binding site.
J.Mol.Biol., 384, 2008
|
|
5DWM
 
 | |
5K0K
 
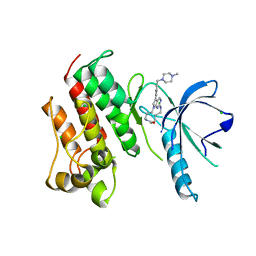 | | Crystal structure of the catalytic domain of the proto-oncogene tyrosine-protein kinase MER in complex with inhibitor UNC2434 | | Descriptor: | 15-{4-[(4-methylpiperazin-1-yl)methyl]phenyl}-4,5,6,7,9,10,11,12-octahydro-2,16-(azenometheno)pyrrolo[2,1-d][1,3,5,9]te traazacyclotetradecin-8(3H)-one, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Wang, X, Liu, J, Zhang, W, Stashko, M.A, Nichols, J, DeRyckere, D, Miley, M.J, Norris-Drouin, J, Chen, Z, Machius, M, Wood, E, Graham, D.K, Earp, H.S, Graham, K, Kireev, D, Frye, S.V. | | Deposit date: | 2016-05-17 | | Release date: | 2017-01-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.545 Å) | | Cite: | Design and Synthesis of Novel Macrocyclic Mer Tyrosine Kinase Inhibitors.
ACS Med Chem Lett, 7, 2016
|
|
5K0X
 
 | | Crystal structure of the catalytic domain of the proto-oncogene tyrosine-protein kinase MER in complex with inhibitor UNC2541 | | Descriptor: | (7S)-7-amino-N-[(4-fluorophenyl)methyl]-8-oxo-2,9,16,18,21-pentaazabicyclo[15.3.1]henicosa-1(21),17,19-triene-20-carboxamide, CHLORIDE ION, Tyrosine-protein kinase Mer | | Authors: | McIver, A.L, Zhang, W, Liu, Q, Jiang, X, Stashko, M.A, Nichols, J, Miley, M.J, Norris-Drouin, J, Machius, M, DeRyckere, D, Wood, E, Graham, D.K, Earp, H.S, Kireev, D, Frye, S.V, Wang, X. | | Deposit date: | 2016-05-17 | | Release date: | 2017-02-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.231 Å) | | Cite: | Discovery of Macrocyclic Pyrimidines as MerTK-Specific Inhibitors.
ChemMedChem, 12, 2017
|
|
6TPX
 
 | |
7QZX
 
 | | Human Carbonic Anhydrase II in complex with 4-oxo-N-(4-sulfamoylphenethyl)-1,3,4,6,7,11b-hexahydro-2H-pyrazino[2,1-a]isoquinoline-2-carboxamide | | Descriptor: | 4-oxo-N-(4-sulfamoylphenethyl)-1,3,4,6,7,11b-hexahydro-2H-pyrazino[2,1-a]isoquinoline-2-carboxamide, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Angeli, A, Ferraroni, M. | | Deposit date: | 2022-02-01 | | Release date: | 2023-02-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Development of Praziquantel sulphonamide derivatives as antischistosomal drugs.
J Enzyme Inhib Med Chem, 37, 2022
|
|
7R1X
 
 | | human Carbonic Anhydrase II in complex with 4-oxo-N-(4-sulfamoylphenethyl)-1,3,4,6,7,11b-hexahydro-2H-pyrazino[2,1-a]isoquinoline-2-carbothioamide | | Descriptor: | 4-oxo-N-(4-sulfamoylphenethyl)-1,3,4,6,7,11b-hexahydro-2H-pyrazino[2,1-a]isoquinoline-2-carbothioamide, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Angeli, A, Ferraroni, M. | | Deposit date: | 2022-02-03 | | Release date: | 2023-02-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Development of Praziquantel sulphonamide derivatives as antischistosomal drugs.
J Enzyme Inhib Med Chem, 37, 2022
|
|
6TLA
 
 | | CRYSTAL STRUCTURE OF LECTIN-LIKE OX-LDL RECEPTOR 1 (C 1 2 1) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Oxidized low-density lipoprotein receptor 1 | | Authors: | Nar, H, Fiegen, D, Schnapp, G. | | Deposit date: | 2019-12-02 | | Release date: | 2020-07-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | A small-molecule inhibitor of lectin-like oxidized LDL receptor-1 acts by stabilizing an inactive receptor tetramer state
Commun Chem, 2020
|
|
6TQ2
 
 | |
6I6K
 
 | | Papaver somniferum O-methyltransferase 1 | | Descriptor: | (13aS)-3,10-dimethoxy-5,8,13,13a-tetrahydro-6H-isoquino[3,2-a]isoquinoline-2,9-diol, O-methyltransferase 1, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Cabry, M.P, Offen, W.A, Winzer, T, Li, Y, Graham, I.A, Davies, G.J, Saleh, P. | | Deposit date: | 2018-11-15 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structure of Papaver somniferum O-Methyltransferase 1 Reveals Initiation of Noscapine Biosynthesis with Implications for Plant Natural Product Methylation
Acs Catalysis, 2019
|
|
4ZR8
 
 | |
6QA8
 
 | | Glycogen Phosphorylase b in complex with 28 | | Descriptor: | (5~{S},7~{R},8~{S},9~{S},10~{R})-7-(hydroxymethyl)-8,9,10-tris(oxidanyl)-2-phenyl-6-oxa-1,3-diazaspiro[4.5]dec-1-en-4-one, Glycogen phosphorylase, muscle form, ... | | Authors: | Kyriakis, E, Stravodimos, G.A, Skamnaki, V.T, Leonidas, D.D. | | Deposit date: | 2018-12-18 | | Release date: | 2019-06-26 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Glucopyranosylidene-spiro-imidazolinones, a New Ring System: Synthesis and Evaluation as Glycogen Phosphorylase Inhibitors by Enzyme Kinetics and X-ray Crystallography.
J.Med.Chem., 62, 2019
|
|
5GXX
 
 | | Crystal structure of endoglucanase CelQ from Clostridium thermocellum complexed with Tris | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Jeng, W.Y, Liu, C.I, Wang, A.H.J. | | Deposit date: | 2016-09-21 | | Release date: | 2017-09-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structures of the C-Terminally Truncated Endoglucanase Cel9Q from Clostridium thermocellum Complexed with Cellodextrins and Tris.
Chembiochem, 20, 2019
|
|
213D
 
 | |
6TNR
 
 | | PI3K delta in complex with N[5(7{2[4(2hydroxypropan2yl)piperidin1 yl]ethoxy}1,3dihydro2benzofuran5yl)2 methoxypyridin3yl]methanesulfonamide | | Descriptor: | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform, ~{N}-[2-methoxy-5-[7-[2-[4-(2-oxidanylpropan-2-yl)piperidin-1-yl]ethoxy]-1,3-dihydro-2-benzofuran-5-yl]pyridin-3-yl]methanesulfonamide | | Authors: | Convery, M.A, Rowland, P, Henley, Z.A, Barton, N, Down, K. | | Deposit date: | 2019-12-10 | | Release date: | 2020-01-01 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Optimization of Orally Bioavailable PI3K delta Inhibitors and Identification of Vps34 as a Key Selectivity Target.
J.Med.Chem., 63, 2020
|
|
5MY6
 
 | | Crystal structure of a HER2-Nb complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Sterckx, Y.G.-J, D'Huyvetter, M, Devoogdt, N. | | Deposit date: | 2017-01-25 | | Release date: | 2017-08-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.246 Å) | | Cite: | (131)I-labeled Anti-HER2 Camelid sdAb as a Theranostic Tool in Cancer Treatment.
Clin. Cancer Res., 23, 2017
|
|
6IAO
 
 | | Structure of Cytochrome P450 BM3 M11 mutant in complex with DTT at resolution 2.16A | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Bifunctional cytochrome P450/NADPH--P450 reductase, CHLORIDE ION, ... | | Authors: | Mirza, O, Rafiq, M, Frydenvang, K. | | Deposit date: | 2018-11-27 | | Release date: | 2019-06-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structural analysis of Cytochrome P450 BM3 mutant M11 in complex with dithiothreitol.
Plos One, 14, 2019
|
|
142D
 
 | |
4CDP
 
 | | Improved coordinates for Escherichia coli O157:H7 heme degrading enzyme ChuS. | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Suits, M.D.L, Jaffer, N, Jia, Z. | | Deposit date: | 2013-11-05 | | Release date: | 2013-11-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of the Escherichia Coli O157:H7 Heme Oxygenase Chus in Complex with Heme and Enzymatic Inactivation by Mutation of the Heme Coordinating Residue His-193.
J.Biol.Chem., 281, 2006
|
|
5K8X
 
 | | Crystal structure of mouse CARM1 in complex with inhibitor U3 | | Descriptor: | 1,2-DIMETHOXYETHANE, 1,2-ETHANEDIOL, 1-METHOXY-2-(2-METHOXYETHOXY)ETHANE, ... | | Authors: | Cura, V, Marechal, N, Mailliot, J, Troffer-Charlier, N, Hassenboehler, P, Wurtz, J.M, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2016-05-31 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.991 Å) | | Cite: | Crystal structure of mouse CARM1 in complex with inhibitor U3
To Be Published
|
|
6EJ3
 
 | | BACE1 compound 23 | | Descriptor: | (1r,4r)-4-methoxy-6'-(5-methyl-3-pyridinyl)-3'H-dispiro[cyclohexane-1,2'-indene-1',4''-[1,3]oxazol]-2''-amine, Beta-secretase 1 | | Authors: | Johansson, P. | | Deposit date: | 2017-09-20 | | Release date: | 2018-04-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Toward beta-Secretase-1 Inhibitors with Improved Isoform Selectivity.
J. Med. Chem., 61, 2018
|
|
6TTI
 
 | | PKM2 in complex with Compound 6 | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, DIMETHYL SULFOXIDE, Pyruvate kinase PKM, ... | | Authors: | Saur, M, Hartshorn, M.J, Dong, J, Reeks, J, Bunkoczi, G, Jhoti, H, Williams, P.A. | | Deposit date: | 2019-12-27 | | Release date: | 2020-01-15 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Fragment-based drug discovery using cryo-EM.
Drug Discov Today, 25, 2020
|
|
8WXL
 
 | | Structure of the SARS-CoV-2 BA.2.86 spike glycoprotein (closed state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yajima, H, Anraku, Y, Kita, S, Kimura, K, Maenaka, K, Hashiguchi, T. | | Deposit date: | 2023-10-30 | | Release date: | 2024-10-09 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | Structural basis for receptor-binding domain mobility of the spike in SARS-CoV-2 BA.2.86 and JN.1.
Nat Commun, 15, 2024
|
|
5JPI
 
 | | 2.15 Angstrom Crystal Structure of S-adenosylhomocysteinase from Cryptosporidium parvum in Complex with D-Eritadenine and NAD | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Adenosylhomocysteinase, ... | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Bishop, B, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-05-03 | | Release date: | 2016-05-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | 2.15 Angstrom Crystal Structure of S-adenosylhomocysteinase from Cryptosporidium parvum in Complex with D-Eritadenine and NAD.
To Be Published
|
|
