8F17
 
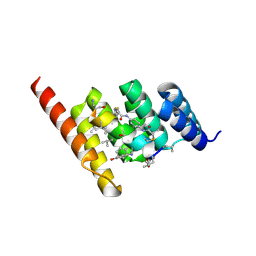 | | Structure of the STUB1 TPR domain in complex with H204, an all-D Helicon Polypeptide | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase CHIP, N,N'-(1,4-phenylene)diacetamide, ... | | Authors: | Li, K, Callahan, A.J, Travaline, T.L, Tokareva, O.S, Swiecicki, J.-M, Verdine, G.L, Pentelute, B.L, McGee, J.H. | | Deposit date: | 2022-11-04 | | Release date: | 2023-02-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Single-Shot Flow Synthesis of D-Proteins for Mirror-Image Phage Display
Chemrxiv, 2023
|
|
5EXM
 
 | |
8F15
 
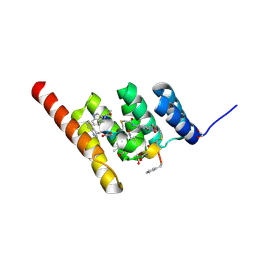 | | Structure of the STUB1 TPR domain in complex with H202, an all-D Helicon Polypeptide | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase CHIP, N,N'-(1,4-phenylene)diacetamide, ... | | Authors: | Li, K, Callahan, A.J, Travaline, T.L, Tokareva, O.S, Swiecicki, J.-M, Verdine, G.L, Pentelute, B.L, McGee, J.H. | | Deposit date: | 2022-11-04 | | Release date: | 2023-02-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Single-Shot Flow Synthesis of D-Proteins for Mirror-Image Phage Display
Chemrxiv, 2023
|
|
2QTU
 
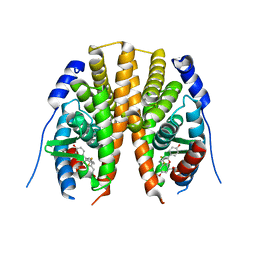 | | Estrogen receptor beta ligand-binding domain complexed to a benzopyran ligand | | Descriptor: | (3aS,4R,9bR)-2,2-difluoro-4-(4-hydroxyphenyl)-6-(methoxymethyl)-1,2,3,3a,4,9b-hexahydrocyclopenta[c]chromen-8-ol, Estrogen receptor beta | | Authors: | Richardson, T.I, Dodge, J.A, Wang, Y, Durbin, J.D, Krishnan, V, Norman, B.H. | | Deposit date: | 2007-08-02 | | Release date: | 2007-10-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Benzopyrans as selective estrogen receptor beta agonists (SERBAs). Part 5: Combined A- and C-ring structure-activity relationship studies.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
8F16
 
 | | Structure of the STUB1 TPR domain in complex with H203, an all-D Helicon Polypeptide | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase CHIP, N,N'-(1,4-phenylene)diacetamide, ... | | Authors: | Li, K, Callahan, A.J, Travaline, T.L, Tokareva, O.S, Swiecicki, J.-M, Verdine, G.L, Pentelute, B.L, McGee, J.H. | | Deposit date: | 2022-11-04 | | Release date: | 2023-02-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Single-Shot Flow Synthesis of D-Proteins for Mirror-Image Phage Display
Chemrxiv, 2023
|
|
8F14
 
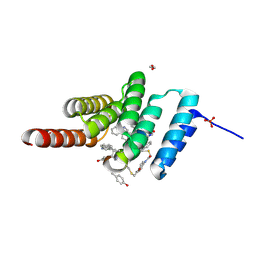 | | Structure of the STUB1 TPR domain in complex with H201, an all-D Helicon Polypeptide | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase CHIP, N,N'-(1,4-phenylene)diacetamide, ... | | Authors: | Li, K, Callahan, A.J, Travaline, T.L, Tokareva, O.S, Swiecicki, J.-M, Verdine, G.L, Pentelute, B.L, McGee, J.H. | | Deposit date: | 2022-11-04 | | Release date: | 2023-02-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Single-Shot Flow Synthesis of D-Proteins for Mirror-Image Phage Display
Chemrxiv, 2023
|
|
6JQM
 
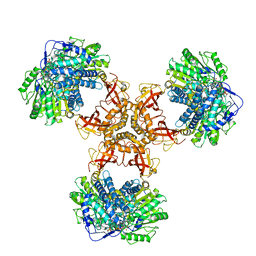 | | Structure of PaaZ with NADPH | | Descriptor: | Bifunctional protein PaaZ, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Gakher, L, Vinothkumar, K.R, Katagihallimath, N, Sowdhamini, R, Sathyanarayanan, N, Cannone, G. | | Deposit date: | 2019-03-31 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular basis for metabolite channeling in a ring opening enzyme of the phenylacetate degradation pathway.
Nat Commun, 10, 2019
|
|
1IYM
 
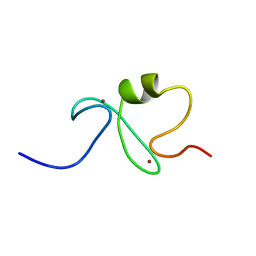 | | RING-H2 finger domain of EL5 | | Descriptor: | EL5, ZINC ION | | Authors: | Katoh, E, Katoh, S, Minami, E, Yamazaki, T. | | Deposit date: | 2002-08-30 | | Release date: | 2003-07-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | High Precision NMR Structure and Function of the RING-H2 Finger Domain of EL5, a Rice Protein Whose Expression Is Increased upon Exposure to Pathogen-derived Oligosaccharides
J.Biol.Chem., 278, 2003
|
|
6V1C
 
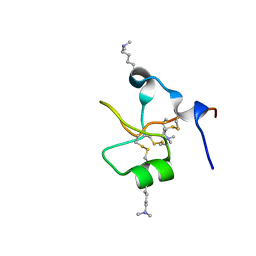 | | Crystal structure of human trefoil factor 3 in complex with its cognate ligand | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-beta-D-galactopyranose, Trefoil factor 3 | | Authors: | Jarva, M.A, Lingford, J.P, John, A, Scott, N.E, Goddard-Borger, E.D. | | Deposit date: | 2019-11-20 | | Release date: | 2019-12-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Trefoil factors share a lectin activity that defines their role in mucus.
Nat Commun, 11, 2020
|
|
4ED5
 
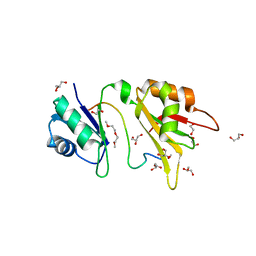 | | Crystal structure of the two N-terminal RRM domains of HuR complexed with RNA | | Descriptor: | 1,2-ETHANEDIOL, 1-METHOXY-2-(2-METHOXYETHOXY)ETHANE, 5'-R(*A*UP*UP*UP*UP*UP*AP*UP*UP*UP*U)-3', ... | | Authors: | Wang, H, Zeng, F, Liu, Q, Niu, L, Teng, M, Li, X. | | Deposit date: | 2012-03-27 | | Release date: | 2012-05-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structure of the ARE-binding domains of Hu antigen R (HuR) undergoes conformational changes during RNA binding.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
1JMG
 
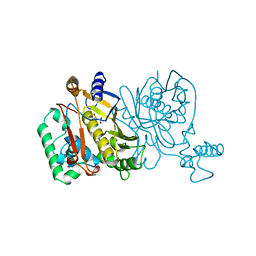 | |
1JEY
 
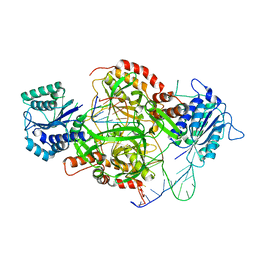 | | Crystal Structure of the Ku heterodimer bound to DNA | | Descriptor: | DNA (34-MER), DNA (5'-D(*GP*TP*TP*TP*TP*TP*AP*GP*TP*TP*TP*AP*TP*TP*GP*GP*GP*CP*GP*CP*G)-3'), Ku70, ... | | Authors: | Walker, J.R, Corpina, R.A, Goldberg, J. | | Deposit date: | 2001-06-19 | | Release date: | 2001-08-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair.
Nature, 412, 2001
|
|
4BAO
 
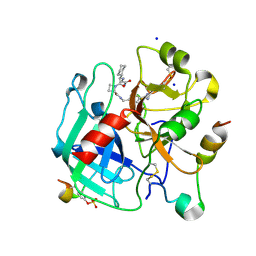 | | Thrombin in complex with inhibitor | | Descriptor: | (2S)-1-[(2R)-2-[(2-azanyl-2-oxidanylidene-ethyl)amino]-2-cyclohexyl-ethanoyl]-N-[(4-carbamimidoylphenyl)methyl]azetidine-2-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HIRUDIN VARIANT-2, ... | | Authors: | Xue, Y, Musil, D. | | Deposit date: | 2012-09-14 | | Release date: | 2013-01-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Identification of Structure-Kinetic and Structure-Thermodynamic Relationships for Thrombin Inhibitors.
Biochemistry, 52, 2013
|
|
6V87
 
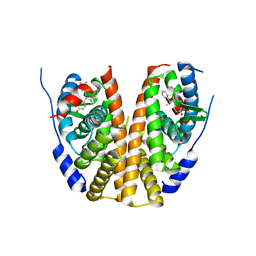 | |
6SWA
 
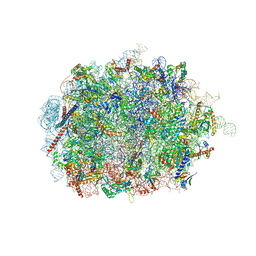 | | Mus musculus brain neocortex ribosome 60S bound to Ebp1 | | Descriptor: | 28S ribosomal RNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Kraushar, M.L, Sprink, T. | | Deposit date: | 2019-09-20 | | Release date: | 2020-09-30 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Protein Synthesis in the Developing Neocortex at Near-Atomic Resolution Reveals Ebp1-Mediated Neuronal Proteostasis at the 60S Tunnel Exit.
Mol.Cell, 81, 2021
|
|
1JCA
 
 | | Non-standard Design of Unstable Insulin Analogues with Enhanced Activity | | Descriptor: | ZINC ION, insulin a, insulin b | | Authors: | Weiss, M.A, Wan, Z, Zhao, M, Chu, Y.-C, Nakagawa, S.H, Burke, G.T, Jia, W, Hellmich, R, Katsoyannis, P.G. | | Deposit date: | 2001-06-08 | | Release date: | 2001-06-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Non-standard insulin design: structure-activity relationships at the periphery of the insulin receptor.
J.Mol.Biol., 315, 2002
|
|
4QB2
 
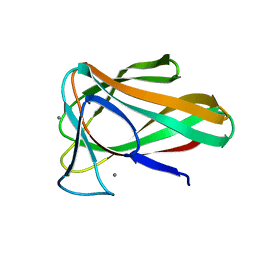 | | Structure of CBM35 in complex with glucuronic acid | | Descriptor: | CALCIUM ION, Xyn30D, alpha-D-glucopyranuronic acid, ... | | Authors: | Sainz-Polo, M.A, Sanz-Aparicio, J. | | Deposit date: | 2014-05-06 | | Release date: | 2014-10-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Analysis of Glucuronoxylan-specific Xyn30D and Its Attached CBM35 Domain Gives Insights into the Role of Modularity in Specificity.
J.Biol.Chem., 289, 2014
|
|
1LO0
 
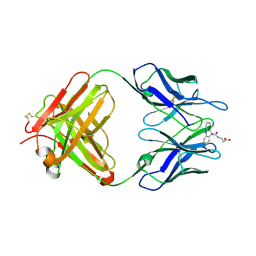 | | Catalytic Retro-Diels-Alderase Transition State Analogue Complex | | Descriptor: | 3-{[(9-CYANO-9,10-DIHYDRO-10-METHYLACRIDIN-9-YL)CARBONYL]AMINO}PROPANOIC ACID, If kappa light chain, Ig gamma 2a heavy chain | | Authors: | Hugot, M, Reymond, J.L, Baumann, U. | | Deposit date: | 2002-05-05 | | Release date: | 2002-06-26 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A structural basis for the activity of retro-Diels-Alder catalytic antibodies: evidence for a catalytic aromatic residue.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1GVH
 
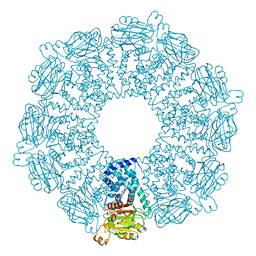 | | The X-ray structure of ferric Escherichia coli flavohemoglobin reveals an unespected geometry of the distal heme pocket | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, FLAVOHEMOPROTEIN, ... | | Authors: | Ilari, A, Johnson, K.A, Bonamore, A, Farina, A, Boffi, A. | | Deposit date: | 2002-02-13 | | Release date: | 2002-08-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | The X-Ray Structure of Ferric Escherichia Coli Flavohemoglobin Reveals an Unexpected Geometry of the Distal Heme Pocket
J.Biol.Chem., 277, 2002
|
|
5FIV
 
 | | STRUCTURAL STUDIES OF HIV AND FIV PROTEASES COMPLEXED WITH AN EFFICIENT INHIBITOR OF FIV PR | | Descriptor: | RETROPEPSIN, benzyl [(1S,4S,7S,8R,9R,10S,13S,16S)-7,10-dibenzyl-8,9-dihydroxy-1,16-dimethyl-4,13-bis(1-methylethyl)-2,5,12,15,18-pentaoxo-20-phenyl-19-oxa-3,6,11,14,17-pentaazaicos-1-yl]carbamate | | Authors: | Li, M, Lee, T, Morris, G, Laco, G, Wong, C, Olson, A, Elder, J, Wlodawer, A, Gustchina, A. | | Deposit date: | 1998-12-02 | | Release date: | 1998-12-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural studies of FIV and HIV-1 proteases complexed with an efficient inhibitor of FIV protease
Proteins, 38, 2000
|
|
5QDW
 
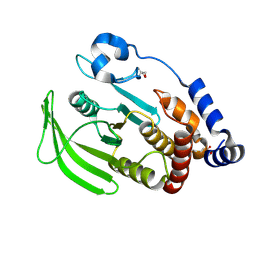 | | PanDDA analysis group deposition -- Crystal structure of PTP1B in complex with compound_FMOPL000465a | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-methoxy-N-[(1R)-1-phenylethyl]acetamide, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Keedy, D.A, Hill, Z.B, Biel, J.T, Kang, E, Rettenmaier, T.J, Brandao-Neto, J, von Delft, F, Wells, J.A, Fraser, J.S. | | Deposit date: | 2018-08-30 | | Release date: | 2018-10-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.212 Å) | | Cite: | An expanded allosteric network in PTP1B by multitemperature crystallography, fragment screening, and covalent tethering.
Elife, 7, 2018
|
|
5QED
 
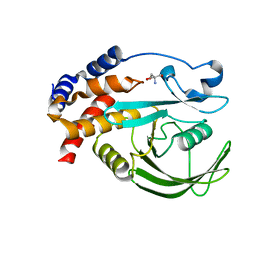 | | PanDDA analysis group deposition -- Crystal structure of PTP1B in complex with compound_FMOPL000538a | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-chloranyl-~{N}-methyl-pyridine-2-carboxamide, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Keedy, D.A, Hill, Z.B, Biel, J.T, Kang, E, Rettenmaier, T.J, Brandao-Neto, J, von Delft, F, Wells, J.A, Fraser, J.S. | | Deposit date: | 2018-08-30 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.755 Å) | | Cite: | An expanded allosteric network in PTP1B by multitemperature crystallography, fragment screening, and covalent tethering.
Elife, 7, 2018
|
|
1LO4
 
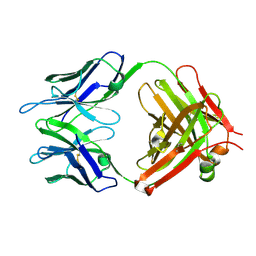 | | Retro-Diels-Alderase Catalytic antibody 9D9 | | Descriptor: | If kappa light chain, Ig gamma 2a heavy chain | | Authors: | Hugot, M, Reymond, J.L, Baumann, U. | | Deposit date: | 2002-05-06 | | Release date: | 2002-07-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A structural basis for the activity of retro-Diels-Alder catalytic antibodies: evidence for a catalytic aromatic residue.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1JQV
 
 | | The K213E mutant of Lactococcus lactis Dihydroorotate dehydrogenase A | | Descriptor: | ACETIC ACID, Dihydroorotate dehydrogenase A, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Norager, S, Arent, S, Bjornberg, O, Ottosen, M, Lo Leggio, L, Jensen, K.F, Larsen, S. | | Deposit date: | 2001-08-09 | | Release date: | 2003-09-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Lactococcus lactis dihydroorotate dehydrogenase A mutants reveal important facets of the enzymatic function
J.Biol.Chem., 278, 2003
|
|
5QDI
 
 | | PanDDA analysis group deposition -- Crystal structure of PTP1B in complex with compound_FMOPL000157a | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, N-(2-phenylethyl)methanesulfonamide, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Keedy, D.A, Hill, Z.B, Biel, J.T, Kang, E, Rettenmaier, T.J, Brandao-Neto, J, von Delft, F, Wells, J.A, Fraser, J.S. | | Deposit date: | 2018-08-30 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.623 Å) | | Cite: | An expanded allosteric network in PTP1B by multitemperature crystallography, fragment screening, and covalent tethering.
Elife, 7, 2018
|
|
