7KND
 
 | | C1B domain of Protein kinase C in apo form | | Descriptor: | Protein kinase C delta type, ZINC ION | | Authors: | Katti, S.S, Krieger, I. | | Deposit date: | 2020-11-04 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Structural anatomy of Protein Kinase C C1 domain interactions with diacylglycerol and other agonists.
Nat Commun, 13, 2022
|
|
7KNJ
 
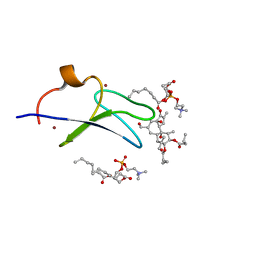 | | C1B domain of Protein kinase C in complex with Phorbol ester and Phosphatidylcholine | | Descriptor: | (4S,7R)-7-(heptanoyloxy)-4-hydroxy-N,N,N-trimethyl-10-oxo-3,5,9-trioxa-4-phosphahexadecan-1-aminium 4-oxide, Phorbol 12,13-dibutyrate, Protein kinase C delta type, ... | | Authors: | Katti, S.S, Krieger, I. | | Deposit date: | 2020-11-04 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structural anatomy of Protein Kinase C C1 domain interactions with diacylglycerol and other agonists.
Nat Commun, 13, 2022
|
|
7KO6
 
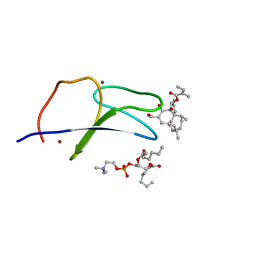 | | C1B domain of Protein kinase C in complex with ingenol-3-angelate and phosphocholine | | Descriptor: | (4S,7R)-7-(heptanoyloxy)-4-hydroxy-N,N,N-trimethyl-10-oxo-3,5,9-trioxa-4-phosphahexadecan-1-aminium 4-oxide, Protein kinase C delta type, ZINC ION, ... | | Authors: | Katti, S.S, Krieger, I.V. | | Deposit date: | 2020-11-06 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural anatomy of Protein Kinase C C1 domain interactions with diacylglycerol and other agonists.
Nat Commun, 13, 2022
|
|
7L92
 
 | |
7LCB
 
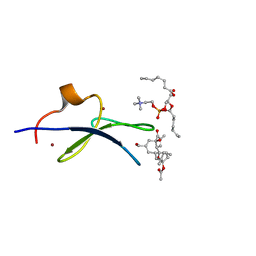 | | C1B domain of Protein kinase C in complex with prostratin and phosphatidylcholine | | Descriptor: | (4S,7R)-7-(heptanoyloxy)-4-hydroxy-N,N,N-trimethyl-10-oxo-3,5,9-trioxa-4-phosphahexadecan-1-aminium 4-oxide, Protein kinase C delta type, ZINC ION, ... | | Authors: | Katti, S.S, Krieger, I.V. | | Deposit date: | 2021-01-10 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural anatomy of Protein Kinase C C1 domain interactions with diacylglycerol and other agonists.
Nat Commun, 13, 2022
|
|
7LEO
 
 | | C1B domain of Protein kinase C in complex with diacylglycerol-lactone (AJH-836) and 1,2-diheptanoyl-sn-glycero-3-phosphocholine | | Descriptor: | (4S,7R)-7-(heptanoyloxy)-4-hydroxy-N,N,N-trimethyl-10-oxo-3,5,9-trioxa-4-phosphahexadecan-1-aminium 4-oxide, Protein kinase C delta type, ZINC ION, ... | | Authors: | Katti, S.S, Krieger, I. | | Deposit date: | 2021-01-14 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural anatomy of Protein Kinase C C1 domain interactions with diacylglycerol and other agonists.
Nat Commun, 13, 2022
|
|
7LF3
 
 | | C1B domain of Protein kinase C in complex with diacylglycerol-lactone AJH-836 | | Descriptor: | (4S,7R)-7-(heptanoyloxy)-4-hydroxy-N,N,N-trimethyl-10-oxo-3,5,9-trioxa-4-phosphahexadecan-1-aminium 4-oxide, Protein kinase C delta type, ZINC ION, ... | | Authors: | Katti, S.S, Krieger, I.V. | | Deposit date: | 2021-01-15 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Structural anatomy of Protein Kinase C C1 domain interactions with diacylglycerol and other agonists.
Nat Commun, 13, 2022
|
|
3KY9
 
 | | Autoinhibited Vav1 | | Descriptor: | Proto-oncogene vav, ZINC ION | | Authors: | Tomchick, D.R, Rosen, M.K, Machius, M, Yu, B. | | Deposit date: | 2009-12-04 | | Release date: | 2010-02-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.731 Å) | | Cite: | Structural and Energetic Mechanisms of Cooperative Autoinhibition and Activation of Vav1
Cell(Cambridge,Mass.), 140, 2010
|
|
5UE8
 
 | | The crystal structure of Munc13-1 C1C2BMUN domain | | Descriptor: | CHLORIDE ION, Protein unc-13 homolog A, ZINC ION | | Authors: | Tomchick, D.R, Rizo, J, Xu, J. | | Deposit date: | 2016-12-29 | | Release date: | 2017-02-15 | | Last modified: | 2018-03-28 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Mechanistic insights into neurotransmitter release and presynaptic plasticity from the crystal structure of Munc13-1 C1C2BMUN.
Elife, 6, 2017
|
|
4L9M
 
 | | Autoinhibited state of the Ras-specific exchange factor RasGRP1 | | Descriptor: | CITRIC ACID, GLYCEROL, RAS guanyl-releasing protein 1, ... | | Authors: | Iwig, J.S, Vercoulen, Y, Das, R, Barros, T, Limnander, A, Che, Y, Pelton, J.G, Wemmer, D.E, Roose, J.P, Kuriyan, J. | | Deposit date: | 2013-06-18 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural analysis of autoinhibition in the Ras-specific exchange factor RasGRP1.
Elife, 2, 2013
|
|
6UAN
 
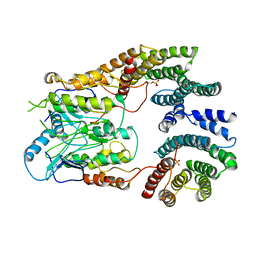 | | B-Raf:14-3-3 complex | | Descriptor: | 14-3-3 zeta, Serine/threonine-protein kinase B-raf | | Authors: | Kondo, Y, Ognjenovic, J, Banerjee, S, Karandur, D, Merk, A, Kulhanek, K, Wong, K, Roose, J.P, Subramaniam, S, Kuriyan, J. | | Deposit date: | 2019-09-11 | | Release date: | 2019-09-25 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structure of a dimeric B-Raf:14-3-3 complex reveals asymmetry in the active sites of B-Raf kinases.
Science, 366, 2019
|
|
6UWA
 
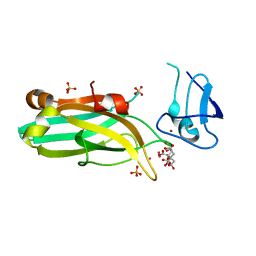 | | Mouse PKC C1B and C2 domains | | Descriptor: | CITRIC ACID, LEAD (II) ION, Protein kinase C, ... | | Authors: | Aggarwal, A, Mire, J, Sacchettini, J.C, Igumenova, T. | | Deposit date: | 2019-11-04 | | Release date: | 2020-12-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Mouse PKC C1B and C2 domains
To Be Published
|
|
7ZR0
 
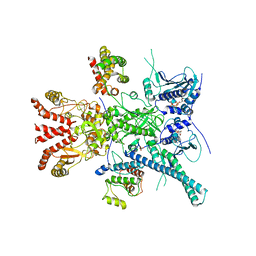 | | CryoEM structure of HSP90-CDC37-BRAF(V600E) complex. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Heat shock protein HSP 90-beta, Hsp90 co-chaperone Cdc37, ... | | Authors: | Oberoi, J, Pearl, L.H. | | Deposit date: | 2022-05-03 | | Release date: | 2022-12-14 | | Last modified: | 2023-01-11 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | HSP90-CDC37-PP5 forms a structural platform for kinase dephosphorylation.
Nat Commun, 13, 2022
|
|
7ZR5
 
 | |
7ZR6
 
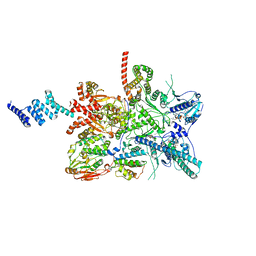 | |
7Z6E
 
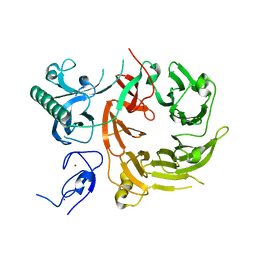 | |
6NFA
 
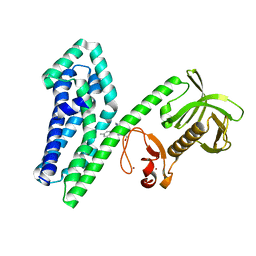 | |
6NEW
 
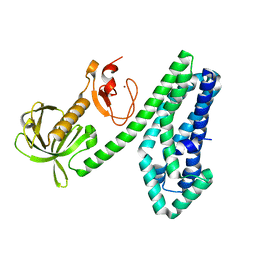 | |
6NF1
 
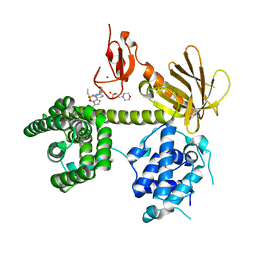 | |
6XHA
 
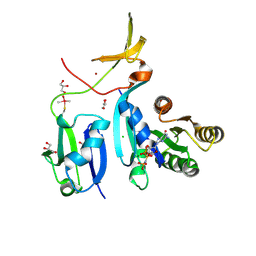 | | Crystal Structure of KRAS-G12V (GMPPNP-bound) in complex with RAS-binding domain (RBD) and cysteine-rich domain (CRD) of RAF1/CRAF | | Descriptor: | CHLORIDE ION, GLYCEROL, Isoform 2B of GTPase KRas, ... | | Authors: | Tran, T.H, Chan, A.H, Dharmaiah, S, Simanshu, D.K. | | Deposit date: | 2020-06-18 | | Release date: | 2021-01-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | KRAS interaction with RAF1 RAS-binding domain and cysteine-rich domain provides insights into RAS-mediated RAF activation.
Nat Commun, 12, 2021
|
|
6XI7
 
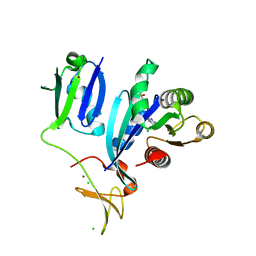 | | Crystal Structure of wild-type KRAS (GMPPNP-bound) in complex with RAS-binding domain (RBD) and cysteine-rich domain (CRD) of RAF1/CRAF (crystal form I) | | Descriptor: | CHLORIDE ION, Isoform 2B of GTPase KRas, MAGNESIUM ION, ... | | Authors: | Chan, A.H, Tran, T.H, Dharmaiah, S, Simanshu, D.K. | | Deposit date: | 2020-06-19 | | Release date: | 2021-01-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | KRAS interaction with RAF1 RAS-binding domain and cysteine-rich domain provides insights into RAS-mediated RAF activation.
Nat Commun, 12, 2021
|
|
7T7R
 
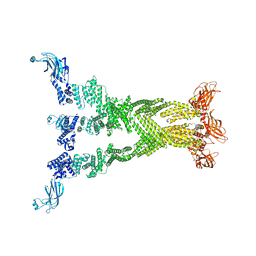 | |
7T7C
 
 | |
7T7V
 
 | |
7T81
 
 | |
