2J9J
 
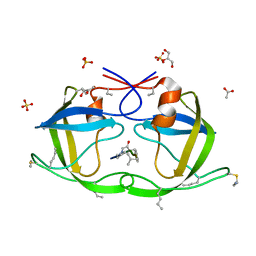 | | Atomic-resolution Crystal Structure of Chemically-Synthesized HIV-1 Protease Complexed with Inhibitor JG-365 | | Descriptor: | ACETATE ION, GLYCEROL, INHIBITOR MOLECULE JG365, ... | | Authors: | Malito, E, Shen, Y, Johnson, E.C.B, Tang, W.J. | | Deposit date: | 2006-11-11 | | Release date: | 2007-08-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Insights from Atomic-Resolution X-Ray Structures of Chemically Synthesized HIV-1 Protease in Complex with Inhibitors.
J.Mol.Biol., 373, 2007
|
|
2JE4
 
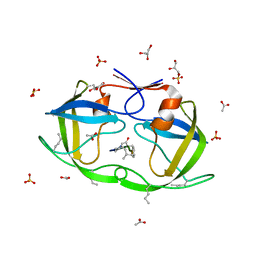 | | Atomic-resolution crystal structure of chemically-synthesized HIV-1 protease in complex with JG-365 | | Descriptor: | ACETATE ION, GLYCEROL, INHIBITOR MOLECULE JG365, ... | | Authors: | Malito, E, Johnson, E.C.B, Tang, W.J. | | Deposit date: | 2007-01-15 | | Release date: | 2007-08-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Modular Total Chemical Synthesis of a Human Immunodeficiency Virus Type 1 Protease.
J.Am.Chem.Soc., 129, 2007
|
|
1KZK
 
 | | JE-2147-HIV Protease Complex | | Descriptor: | (4R)-3-{(2S,3S)-2-hydroxy-3-[(3-hydroxy-2-methylbenzoyl)amino]-4-phenylbutanoyl}-5,5-dimethyl-N-(2-methylbenzyl)-1,3-thiazolidine-4-carboxamide, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Reiling, K.K, Endres, N.F, Dauber, D.S, Craik, C.S, Stroud, R.M. | | Deposit date: | 2002-02-06 | | Release date: | 2002-04-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Anisotropic Dynamics of the JE-2147-HIV Protease Complex:
Drug Resistance and Thermodynamic Binding Mode Examined in a 1.09 A Structure
Biochemistry, 41, 2002
|
|
1T3R
 
 | | HIV protease wild-type in complex with TMC114 inhibitor | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, PHOSPHATE ION, protease RETROPEPSIN | | Authors: | King, N.M, Prabu-Jeyabalan, M, Nalivaika, E.A, Wigernick, P.B, de Bethune, M.P, Schiffer, C.A. | | Deposit date: | 2004-04-27 | | Release date: | 2005-05-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Discovery and selection of TMC114, a next generation HIV-1 protease inhibitor
J.Med.Chem., 48, 2005
|
|
2J9K
 
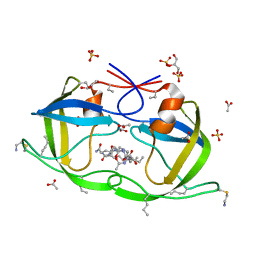 | | Atomic-resolution Crystal Structure of Chemically-Synthesized HIV-1 Protease Complexed with Inhibitor MVT-101 | | Descriptor: | ACETATE ION, GLYCEROL, N-{(2S)-2-[(N-acetyl-L-threonyl-L-isoleucyl)amino]hexyl}-L-norleucyl-L-glutaminyl-N~5~-[amino(iminio)methyl]-L-ornithinamide, ... | | Authors: | Malito, E, Shen, Y, Johnson, E.C.B, Tang, W.J. | | Deposit date: | 2006-11-11 | | Release date: | 2007-08-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Insights from Atomic-Resolution X-Ray Structures of Chemically Synthesized HIV-1 Protease in Complex with Inhibitors.
J.Mol.Biol., 373, 2007
|
|
3HAU
 
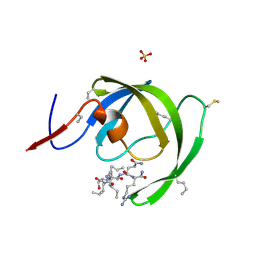 | |
3HAW
 
 | |
4DJQ
 
 | | Crystal Structure of wild-type HIV-1 Protease in Complex with MKP86 | | Descriptor: | 2-(2-oxoimidazolidin-1-yl)ethyl [(2S,3R)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl][(2S)-2-methylbutyl]amino}-1-phenylbutan-2-yl]carbamate, PHOSPHATE ION, Pol polyprotein | | Authors: | Schiffer, C.A, Nalam, M.N.L. | | Deposit date: | 2012-02-02 | | Release date: | 2012-08-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Design, synthesis, and biological and structural evaluations of novel HIV-1 protease inhibitors to combat drug resistance.
J.Med.Chem., 55, 2012
|
|
3KA2
 
 | |
4DJP
 
 | | Crystal Structure of wild-type HIV-1 Protease in Complex with MKP73 | | Descriptor: | PHOSPHATE ION, Pol polyprotein, methyl (2S)-3-({[(2S,3R)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl][(2S)-2-methylbutyl]amino}-1-phenylbutan-2-yl]carbamoyl}oxy)-2-methylpropanoate | | Authors: | Schiffer, C.A, Nalam, M.N.L. | | Deposit date: | 2012-02-02 | | Release date: | 2012-08-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Design, synthesis, and biological and structural evaluations of novel HIV-1 protease inhibitors to combat drug resistance.
J.Med.Chem., 55, 2012
|
|
2FNT
 
 | | Crystal structure of a drug-resistant (V82A) inactive (D25N) HIV-1 protease complexed with AP2V variant of HIV-1 NC-p1 substrate. | | Descriptor: | ACETATE ION, NC-p1 substrate PEPTIDE, PHOSPHATE ION, ... | | Authors: | Prabu-Jeyabaln, M, Nalivaika, E.A, Schiffer, C.A. | | Deposit date: | 2006-01-11 | | Release date: | 2006-09-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Mechanism of substrate recognition by drug-resistant human immunodeficiency virus type 1 protease variants revealed by a novel structural intermediate.
J.Virol., 80, 2006
|
|
3NYG
 
 | |
3O9I
 
 | | Crystal Structure of wild-type HIV-1 Protease in complex with af61 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl {(1S,2R)-3-[(1,3-benzothiazol-6-ylsulfonyl)(2-ethylbutyl)amino]-1-benzyl-2-hydroxypropyl}carbamate, PHOSPHATE ION, Protease | | Authors: | Schiffer, C.A, Nalam, M.N.L. | | Deposit date: | 2010-08-04 | | Release date: | 2011-08-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Substrate envelope-designed potent HIV-1 protease inhibitors to avoid drug resistance.
Chem.Biol., 20, 2013
|
|
3O9E
 
 | | Crystal Structure of wild-type HIV-1 Protease in complex with af60 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(1S,2R)-3-{[(4-aminophenyl)sulfonyl](2-ethylbutyl)amino}-1-benzyl-2-hydroxypropyl]carbamate, ACETATE ION, PHOSPHATE ION, ... | | Authors: | Schiffer, C.A, Nalam, M.N.L. | | Deposit date: | 2010-08-04 | | Release date: | 2011-08-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Substrate envelope-designed potent HIV-1 protease inhibitors to avoid drug resistance.
Chem.Biol., 20, 2013
|
|
2FGV
 
 | | X-ray crystal structure of HIV-1 Protease T80N variant in complex with the inhibitor saquinavir used to explore the role of invariant Thr80 in HIV-1 protease structure, function, and viral infectivity. | | Descriptor: | (2S)-2-amino-3-phenylpropane-1,1-diol, 2-METHYL-DECAHYDRO-ISOQUINOLINE-3-CARBOXYLIC ACID, ASPARAGINE, ... | | Authors: | Foulkes, J.E, Prabu-Jeyabalan, M, Cooper, D, Schiffer, C.A. | | Deposit date: | 2005-12-22 | | Release date: | 2006-11-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Role of invariant Thr80 in human immunodeficiency virus type 1 protease structure, function, and viral infectivity.
J.Virol., 80, 2006
|
|
3NWQ
 
 | |
4QJ6
 
 | |
3O9B
 
 | | Crystal Structure of wild-type HIV-1 Protease in Complex with kd25 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(1S,2R)-1-benzyl-2-hydroxy-3-({[4-(hydroxymethyl)phenyl]sulfonyl}[(2S)-2-methylbutyl]amino)propyl]carbamate, ACETATE ION, GLYCEROL, ... | | Authors: | Schiffer, C.A, Nalam, M.N.L. | | Deposit date: | 2010-08-04 | | Release date: | 2011-08-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Substrate envelope-designed potent HIV-1 protease inhibitors to avoid drug resistance.
Chem.Biol., 20, 2013
|
|
4QJA
 
 | |
4DJR
 
 | | Crystal Structure of wild-type HIV-1 Protease in Complex with MKP97 | | Descriptor: | PHOSPHATE ION, Pol polyprotein, [(2S)-5-oxopyrrolidin-2-yl]methyl [(2S,3R)-4-{(1,3-benzothiazol-6-ylsulfonyl)[(2S)-2-methylbutyl]amino}-3-hydroxy-1-phenylbutan-2-yl]carbamate | | Authors: | Schiffer, C.A, Nalam, M.N.L. | | Deposit date: | 2012-02-02 | | Release date: | 2012-08-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Design, synthesis, and biological and structural evaluations of novel HIV-1 protease inhibitors to combat drug resistance.
J.Med.Chem., 55, 2012
|
|
2F3K
 
 | | Substrate envelope and drug resistance: crystal structure of r01 in complex with wild-type hiv-1 protease | | Descriptor: | (3S,4AS,8AS)-N-(TERT-BUTYL)-2-[(3S)-3-({3-(METHYLSULFONYL)-N-[(PYRIDIN-3-YLOXY)ACETYL]-L-VALYL}AMINO)-2-OXO-4-PHENYLBUTYL]DECAHYDROISOQUINOLINE-3-CARBOXAMIDE, PHOSPHATE ION, Protease | | Authors: | Prabu-Jeyabalan, M, King, N.M, Nalivaika, E.A, Heilek-Snyder, G, Cammack, N, Schiffer, C.A. | | Deposit date: | 2005-11-21 | | Release date: | 2006-11-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Substrate envelope and drug resistance: crystal structure of RO1 in complex with wild-type human immunodeficiency virus type 1 protease.
Antimicrob.Agents Chemother., 50, 2006
|
|
3BXR
 
 | | Crystal Structures Of Highly Constrained Substrate And Hydrolysis Products Bound To HIV-1 Protease. Implications For Catalytic Mechanism | | Descriptor: | (9S,12S)-9-(1-methylethyl)-N-[(8S,11S)-8-[(1S)-1-methylpropyl]-7,10-dioxo-2-oxa-6,9-diazabicyclo[11.2.2]heptadeca-1(15),13,16-trien-11-yl]-7,10-dioxo-2-oxa-8,11-diazabicyclo[12.2.2]octadeca-1(16),14,17-triene-12-carboxamide, Protease, SULFATE ION | | Authors: | Tyndall, J.D, Pattenden, L.K, Reid, R.C, Hu, S.H, Alewood, D, Alewood, P.F, Walsh, T, Fairlie, D.P, Martin, J.L. | | Deposit date: | 2008-01-14 | | Release date: | 2008-03-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structures of Highly Constrained Substrate and Hydrolysis Products Bound to HIV-1 Protease. Implications for the Catalytic Mechanism
Biochemistry, 47, 2008
|
|
3HLO
 
 | |
3FSM
 
 | |
3EL5
 
 | |
