9UJ2
 
 | | 14-3-3 zeta chimera with the S202R peptide of SARS-CoV-2 N (residues 200-213) | | Descriptor: | 1,2-ETHANEDIOL, 14-3-3 protein zeta/delta,Peptide from Nucleoprotein, DI(HYDROXYETHYL)ETHER | | Authors: | Boyko, K.M, Matyuta, I.O, Minyaev, M.E, Perfilova, K.V, Sluchanko, N.N. | | Deposit date: | 2025-04-16 | | Release date: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High-resolution structure reveals enhanced 14-3-3 binding by a mutant SARS-CoV-2 nucleoprotein variant with improved replicative fitness.
Biochem.Biophys.Res.Commun., 767, 2025
|
|
4Y5I
 
 | | Crystal structure of C-terminal modified Tau peptide-hybrid 126B with 14-3-3sigma | | Descriptor: | 14-3-3 protein sigma, CHLORIDE ION, Microtubule-associated protein tau | | Authors: | Leysen, S, Bartel, M, Milroy, L, Brunsveld, L, Ottmann, C. | | Deposit date: | 2015-02-11 | | Release date: | 2016-01-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Stabilizer-Guided Inhibition of Protein-Protein Interactions.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
1YZ5
 
 | |
5XY9
 
 | | Structure of the MST4 and 14-3-3 complex | | Descriptor: | 14-3-3 protein zeta/delta, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, GLYCEROL, ... | | Authors: | Shi, Z.B, Zhou, Z.C. | | Deposit date: | 2017-07-06 | | Release date: | 2018-07-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | Structure of the MST4 and 14-3-3 complex
To Be Published
|
|
7EXE
 
 | |
8HEW
 
 | | Potato 14-3-3 St14f | | Descriptor: | 14-3-3 protein, StFDL1 peptide | | Authors: | Taoka, K, Kawahara, I, Shinya, S, Harada, K, Muranaka, T, Furuita, K, Nakagawa, A, Fujiwara, T, Tsuji, H, Kojima, C. | | Deposit date: | 2022-11-08 | | Release date: | 2023-09-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Multifunctional chemical inhibitors of the florigen activation complex discovered by structure-based high-throughput screening.
Plant J., 112, 2022
|
|
1YWT
 
 | | Crystal structure of the human sigma isoform of 14-3-3 in complex with a mode-1 phosphopeptide | | Descriptor: | 14-3-3 protein sigma, CALCIUM ION, synthetic optimal phosphopeptide (mode-1) | | Authors: | Wilker, E.W, Grant, R.A, Artim, S.C, Yaffe, M.B. | | Deposit date: | 2005-02-18 | | Release date: | 2005-03-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A structural basis for 14-3-3sigma functional specificity.
J.Biol.Chem., 280, 2005
|
|
4N7G
 
 | | Crystal structure of 14-3-3zeta in complex with a peptide derived from ExoS | | Descriptor: | 14-3-3 protein zeta/delta, Exoenzyme S, HEXAETHYLENE GLYCOL | | Authors: | Bier, D, Glas, A, Hahne, G, Grossmann, T, Ottmann, C. | | Deposit date: | 2013-10-15 | | Release date: | 2014-02-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Constrained peptides with target-adapted cross-links as inhibitors of a pathogenic protein-protein interaction.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
1QJA
 
 | | 14-3-3 ZETA/PHOSPHOPEPTIDE COMPLEX (MODE 2) | | Descriptor: | 14-3-3 PROTEIN ZETA, PHOSPHOPEPTIDE | | Authors: | Rittinger, K, Budman, J, Xu, J, Volinia, S, Cantley, L.C, Smerdon, S.J, Gamblin, S.J, Yaffe, M.B. | | Deposit date: | 1999-06-23 | | Release date: | 1999-09-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Analysis of 14-3-3 Phosphopeptide Complexes Identifies a Dual Role for the Nuclear Export Signal of 14-3-3 in Ligand Binding
Mol.Cell, 4, 1999
|
|
4N84
 
 | | Crystal structure of 14-3-3zeta in complex with a 12-carbon-linker cyclic peptide derived from ExoS | | Descriptor: | 14-3-3 protein zeta/delta, Exoenzyme S | | Authors: | Bier, D, Glas, A, Hahne, G, Grossmann, T, Ottmann, C. | | Deposit date: | 2013-10-17 | | Release date: | 2014-02-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Constrained peptides with target-adapted cross-links as inhibitors of a pathogenic protein-protein interaction.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
7V9B
 
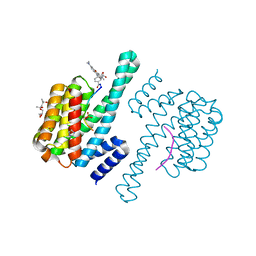 | | Crystal Structure of 14-3-3 epsilon with FOXO3a peptide | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-[3,6-bis(dimethylamino)xanthen-9-yl]-5-methanoyl-benzoate, ARG-ARG-ARG-ALA-VAL-SEP-MET-ASP-ASN-SER-ASN, ... | | Authors: | Mathivanan, S, Kamariah, N. | | Deposit date: | 2021-08-24 | | Release date: | 2022-08-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of a 14-3-3 epsilon :FOXO3a pS253 Phosphopeptide Complex Reveals 14-3-3 Isoform-Specific Binding of Forkhead Box Class O Transcription Factor (FOXO) Phosphoproteins.
Acs Omega, 7, 2022
|
|
1QJB
 
 | | 14-3-3 ZETA/PHOSPHOPEPTIDE COMPLEX (MODE 1) | | Descriptor: | 14-3-3 PROTEIN ZETA/DELTA, PHOSPHOPEPTIDE | | Authors: | Rittinger, K, Budman, J, Xu, J, Volinia, S, Cantley, L.C, Smerdon, S.J, Gamblin, S.J, Yaffe, M.B. | | Deposit date: | 1999-06-23 | | Release date: | 1999-09-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Analysis of 14-3-3 Phosphopeptide Complexes Identifies a Dual Role for the Nuclear Export Signal of 14-3-3 in Ligand Binding
Mol.Cell, 4, 1999
|
|
6NV2
 
 | | 14-3-3 sigma with RelA/p65 binding site pS45 in complex with DP005 | | Descriptor: | (4R,5R,6R,6aS,9S,9aE,10aR)-5-hydroxy-9-(methoxymethyl)-6,10a-dimethyl-3-(propan-2-yl)-1,2,4,5,6,6a,7,8,9,10a-decahydrodicyclopenta[a,d][8]annulen-4-yl alpha-D-glucopyranoside, 14-3-3 protein sigma, CHLORIDE ION, ... | | Authors: | Wolter, M, Ottmann, C. | | Deposit date: | 2019-02-04 | | Release date: | 2020-05-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Selectivity via Cooperativity: Preferential Stabilization of the p65/14-3-3 Interaction with Semisynthetic Natural Products.
J.Am.Chem.Soc., 142, 2020
|
|
2V7D
 
 | |
6KZH
 
 | |
8P1H
 
 | | Crystal structure of the chimera of human 14-3-3 zeta and phosphorylated cytoplasmic loop fragment of the alpha7 acetylcholine receptor | | Descriptor: | 1,2-ETHANEDIOL, AZIDE ION, BENZOIC ACID, ... | | Authors: | Boyko, K.M, Kapitonova, A.A, Tugaeva, K.V, Varfolomeeva, L.A, Lyukmanova, E.N, Sluchanko, N.N. | | Deposit date: | 2023-05-12 | | Release date: | 2023-10-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure reveals canonical recognition of the phosphorylated cytoplasmic loop of human alpha7 nicotinic acetylcholine receptor by 14-3-3 protein.
Biochem.Biophys.Res.Commun., 682, 2023
|
|
6KZG
 
 | |
8P0D
 
 | | Human 14-3-3 sigma in complex with human MDM2 peptide | | Descriptor: | 14-3-3 protein sigma, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Roversi, P, Ward, J, Doveston, R, Kwon, H, Romartinez Alonso, B. | | Deposit date: | 2023-05-10 | | Release date: | 2024-02-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Characterizing the protein-protein interaction between MDM2 and 14-3-3 sigma ; proof of concept for small molecule stabilization.
J.Biol.Chem., 300, 2024
|
|
7XBQ
 
 | |
2WH0
 
 | | Recognition of an intrachain tandem 14-3-3 binding site within protein kinase C epsilon | | Descriptor: | 14-3-3 PROTEIN ZETA/DELTA, CALCIUM ION, PROTEIN KINASE C EPSILON TYPE, ... | | Authors: | Kostelecky, B, Saurin, A.T, Purkiss, A, Parker, P.J, McDonald, N.Q. | | Deposit date: | 2009-04-28 | | Release date: | 2009-08-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Recognition of an Intra-Chain Tandem 14-3-3 Binding Site within Pkc Epsilon.
Embo Rep., 10, 2009
|
|
4DX0
 
 | | Structure of the 14-3-3/PMA2 complex stabilized by a pyrazole derivative | | Descriptor: | 14-3-3-like protein E, 4-[(4R)-4-(4-nitrophenyl)-6-oxidanylidene-3-phenyl-1,4-dihydropyrrolo[3,4-c]pyrazol-5-yl]benzoic acid, N.plumbaginifolia H+-translocating ATPase mRNA | | Authors: | Richter, A, Rose, R, Hedberg, C, Waldmann, H, Ottmann, C. | | Deposit date: | 2012-02-27 | | Release date: | 2012-05-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | An Optimised Small-Molecule Stabiliser of the 14-3-3-PMA2 Protein-Protein Interaction.
Chemistry, 18, 2012
|
|
3T0M
 
 | |
4DHP
 
 | | Small-molecule inhibitors of 14-3-3 protein-protein interactions from virtual screening | | Descriptor: | 14-3-3 protein sigma, CHLORIDE ION, GLYCEROL, ... | | Authors: | Thiel, P, Roeglin, L, Kohlbacher, O, Ottmann, C. | | Deposit date: | 2012-01-30 | | Release date: | 2013-07-31 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Virtual screening and experimental validation reveal novel small-molecule inhibitors of 14-3-3 protein-protein interactions.
Chem.Commun.(Camb.), 49, 2013
|
|
3SP5
 
 | | Crystal structure of human 14-3-3 sigma C38V/N166H in complex with TASK-3 peptide and stabilizer Cotylenol | | Descriptor: | 14-3-3 protein sigma, Cotylenol, MAGNESIUM ION, ... | | Authors: | Anders, C, Schumacher, B, Ottmann, C. | | Deposit date: | 2011-07-01 | | Release date: | 2016-05-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A semisynthetic fusicoccane stabilizes a protein-protein interaction and enhances the expression of K+ channels at the cell surface.
Chem.Biol., 20, 2013
|
|
3SPR
 
 | | Crystal structure of human 14-3-3 sigma C38V/N166H in complex with TASK-3 peptide and stabilizer FC-THF | | Descriptor: | 14-3-3 protein sigma, CHLORIDE ION, Fusicoccin A-THF, ... | | Authors: | Anders, C, Schumacher, B, Ottmann, C. | | Deposit date: | 2011-07-03 | | Release date: | 2012-07-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | A semisynthetic fusicoccane stabilizes a protein-protein interaction and enhances the expression of K+ channels at the cell surface.
Chem. Biol., 20, 2013
|
|
