6TL7
 
 | |
2LS8
 
 | | Solution structure of human C-type lectin domain family 4 member D | | Descriptor: | C-type lectin domain family 4 member D | | Authors: | Harris, R, Gaudette, J, Bandaranayake, A.D, Banu, R, Bonanno, J.B, Calarese, D.A, Celikgil, A, Chamala, S, Chan, M.K, Chaparro, R, Evans, B, Garforth, S, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Lim, S, Love, J, Matikainen, B, Patel, H, Seidel, R.D, Smith, B, Stead, M, Girvin, M.E, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-04-23 | | Release date: | 2012-05-09 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of human C-type lectin domain family 4 member D
To be Published
|
|
4J6K
 
 | |
7ODU
 
 | | Natural killer cell receptor NKR-P1B from Rattus norvegicus in complex with its cognate ligand Clr-11 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, C-type lectin domain family 2 member D11, ... | | Authors: | Skalova, T, Blaha, J, Kalouskova, B, Skorepa, O, Vanek, O, Dohnalek, J. | | Deposit date: | 2021-04-30 | | Release date: | 2022-06-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Natural killer cell receptor NKR-P1B from Rattus norvegicus in complex with its cognate ligand Clr-11
To Be Published
|
|
6GHV
 
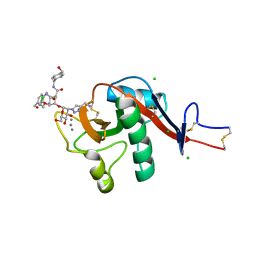 | | Structure of a DC-SIGN CRD in complex with high affinity glycomimetic. | | Descriptor: | CALCIUM ION, CD209 antigen, CHLORIDE ION, ... | | Authors: | Thepaut, M, Achilli, S, Medve, L, Bernardi, A, Fieschi, F. | | Deposit date: | 2018-05-09 | | Release date: | 2019-09-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Enhancing Potency and Selectivity of a DC-SIGN Glycomimetic Ligand by Fragment-Based Design: Structural Basis.
Chemistry, 25, 2019
|
|
6W12
 
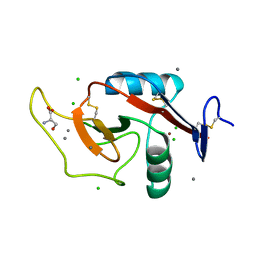 | | Crystal Structure of the Carbohydrate Recognition Domain of the Human Macrophage Galactose C-Type Lectin Bound to the Tumor-Associated Tn Antigen | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, C-type lectin domain family 10 member A, CALCIUM ION, ... | | Authors: | Birrane, G, Murphy, P.V, Gabba, A, Luz, J.G. | | Deposit date: | 2020-03-03 | | Release date: | 2021-03-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Carbohydrate Recognition Domain of the Human Macrophage Galactose C-Type Lectin Bound to GalNAc and the Tumor-Associated Tn Antigen.
Biochemistry, 60, 2021
|
|
8A59
 
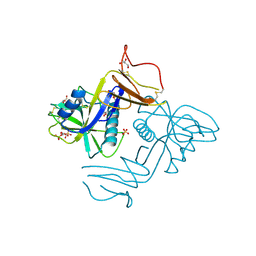 | | C-type lectin-like domain (CTLD) and Sushi-like domain of human CD93 | | Descriptor: | Complement component C1q receptor, GLYCEROL, SULFATE ION | | Authors: | Tassone, G, Barbera, S, Raucci, L, Orlandini, M, Pozzi, C. | | Deposit date: | 2022-06-14 | | Release date: | 2022-11-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Dimerization of the C-type lectin-like receptor CD93 promotes its binding to Multimerin-2 in endothelial cells.
Int.J.Biol.Macromol., 224, 2023
|
|
7W5D
 
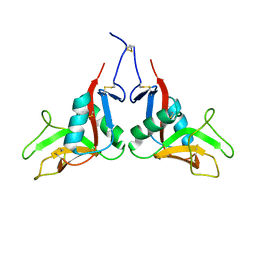 | |
6E7D
 
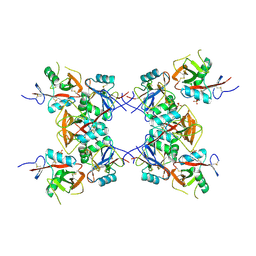 | | Structure of the inhibitory NKR-P1B receptor bound to the host-encoded ligand, Clr-b | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C-type lectin domain family 2 member D, Killer cell lectin-like receptor subfamily B member 1B allele B, ... | | Authors: | Balaji, G.R, Rossjohn, J, Berry, R. | | Deposit date: | 2018-07-26 | | Release date: | 2018-10-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Recognition of host Clr-b by the inhibitory NKR-P1B receptor provides a basis for missing-self recognition.
Nat Commun, 9, 2018
|
|
8TS0
 
 | |
1MSB
 
 | |
3KQG
 
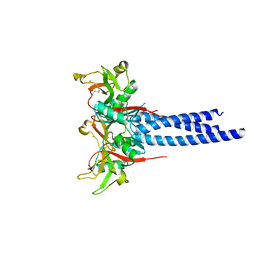 | | Trimeric Structure of Langerin | | Descriptor: | C-type lectin domain family 4 member K, CALCIUM ION | | Authors: | Feinberg, H, Powlesland, A.S, Taylor, M.E, Weis, W.I. | | Deposit date: | 2009-11-17 | | Release date: | 2010-02-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Trimeric structure of langerin.
J.Biol.Chem., 285, 2010
|
|
3KMB
 
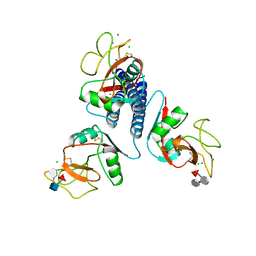 | |
1MUQ
 
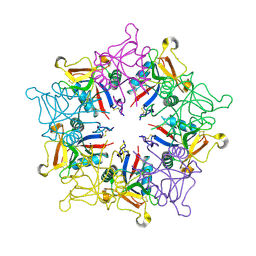 | | X-ray Crystal Structure of Rattlesnake Venom Complexed With Thiodigalactoside | | Descriptor: | 1-thio-beta-D-galactopyranose-(1-1)-beta-D-galactopyranose, CALCIUM ION, Galactose-specific lectin, ... | | Authors: | Walker, J.R, Nagar, B, Young, N.M, Hirama, T, Rini, J.M. | | Deposit date: | 2002-09-24 | | Release date: | 2003-07-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray Crystal Structure of a Galactose-Specific C-Type Lectin Possessing a Novel Decameric Quaternary Structure.
Biochemistry, 43, 2004
|
|
5VYB
 
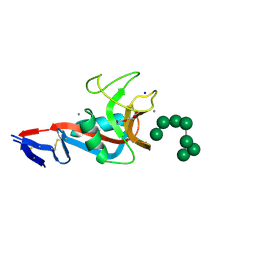 | | Structure of the carbohydrate recognition domain of Dectin-2 complexed with a mammalian-type high mannose Man9GlcNAc2 oligosaccharide | | Descriptor: | C-type lectin domain family 6 member A, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Feinberg, H, Jegouzo, S.A.F, Rex, M.J, Drickamer, K, Taylor, M.E, Weis, W.I. | | Deposit date: | 2017-05-24 | | Release date: | 2017-07-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mechanism of pathogen recognition by human dectin-2.
J. Biol. Chem., 292, 2017
|
|
7NL7
 
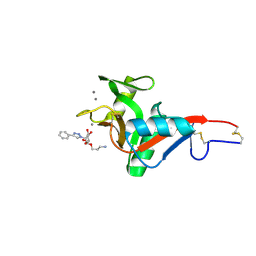 | | Crystal Structure of DC-SIGN in complex with a triazole-based glycomimetic ligand | | Descriptor: | 3-Aminopropyl 2-deoxy-2-(4-phenyl-1,2,3-triazol-1-yl)-alpha-D-mannopyranoside, CALCIUM ION, DC-SIGN, ... | | Authors: | Jakob, R.P, Cramer, J, Lakkaichi, A, Aliu, B, Cattaneo, I, Klein, S, Jiang, X, Rabbani, S, Schwardt, O, Ernst, B, Maier, T. | | Deposit date: | 2021-02-22 | | Release date: | 2021-10-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Sweet Drugs for Bad Bugs: A Glycomimetic Strategy against the DC-SIGN-Mediated Dissemination of SARS-CoV-2.
J.Am.Chem.Soc., 143, 2021
|
|
7NL6
 
 | | Crystal Structure of DC-SIGN in complex with a triazole-based glycomimetic ligand | | Descriptor: | CALCIUM ION, DC-SIGN, CRD domain, ... | | Authors: | Jakob, R.P, Cramer, J, Lakkaichi, A, Aliu, B, Cattaneo, I, Klein, S, Jiang, X, Rabbani, S, Schwardt, O, Ernst, B, Maier, T. | | Deposit date: | 2021-02-22 | | Release date: | 2021-10-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Sweet Drugs for Bad Bugs: A Glycomimetic Strategy against the DC-SIGN-Mediated Dissemination of SARS-CoV-2.
J.Am.Chem.Soc., 143, 2021
|
|
5MGR
 
 | | Human receptor NKR-P1 in glycosylated form, extracellular domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Killer cell lectin-like receptor subfamily B member 1, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Skalova, T, Blaha, J, Stransky, J, Koval, T, Hasek, J, Yuguang, Z, Harlos, K, Vanek, O, Dohnalek, J. | | Deposit date: | 2016-11-22 | | Release date: | 2018-06-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the human NK cell NKR-P1:LLT1 receptor:ligand complex reveals clustering in the immune synapse.
Nat Commun, 13, 2022
|
|
8URF
 
 | |
5G6U
 
 | | Crystal structure of langerin carbohydrate recognition domain with GlcNS6S | | Descriptor: | 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Porkolab, V, Chabrol, E, Varga, N, Ordanini, S, Sutkeviciute, I, Thepaut, M, Bernardi, A, Fieschi, F. | | Deposit date: | 2016-07-21 | | Release date: | 2018-02-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.844 Å) | | Cite: | Rational-Differential Design of Highly Specific Glycomimetic Ligands: Targeting DC-SIGN and Excluding Langerin Recognition.
ACS Chem. Biol., 13, 2018
|
|
5M62
 
 | | Structure of the Mus musclus Langerin carbohydrate recognition domain in complex with glucose | | Descriptor: | C-type lectin domain family 4 member K, CALCIUM ION, GLYCEROL, ... | | Authors: | Loll, B, Aretz, J, Rademacher, C, Wahl, M.C. | | Deposit date: | 2016-10-24 | | Release date: | 2016-12-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Bacterial Polysaccharide Specificity of the Pattern Recognition Receptor Langerin Is Highly Species-dependent.
J. Biol. Chem., 292, 2017
|
|
1EGG
 
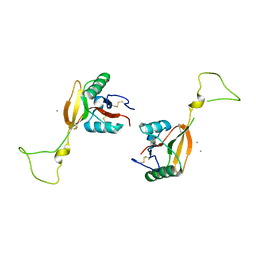 | | STRUCTURE OF A C-TYPE CARBOHYDRATE-RECOGNITION DOMAIN (CRD-4) FROM THE MACROPHAGE MANNOSE RECEPTOR | | Descriptor: | CALCIUM ION, MACROPHAGE MANNOSE RECEPTOR | | Authors: | Feinberg, H, Park-Snyder, S, Kolatkar, A.R, Heise, C.T, Taylor, M.E, Weis, W.I. | | Deposit date: | 2000-02-15 | | Release date: | 2000-08-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of a C-type carbohydrate recognition domain from the macrophage mannose receptor.
J.Biol.Chem., 275, 2000
|
|
5MGS
 
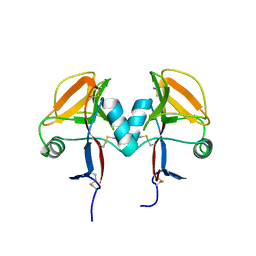 | | Human receptor NKR-P1 in deglycosylated form, extracellular domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Killer cell lectin-like receptor subfamily B member 1 | | Authors: | Skalova, T, Blaha, J, Stransky, J, Koval, T, Hasek, J, Yuguang, Z, Harlos, K, Vanek, O, Dohnalek, J. | | Deposit date: | 2016-11-22 | | Release date: | 2018-05-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the human NK cell NKR-P1:LLT1 receptor:ligand complex reveals clustering in the immune synapse.
Nat Commun, 13, 2022
|
|
1E8I
 
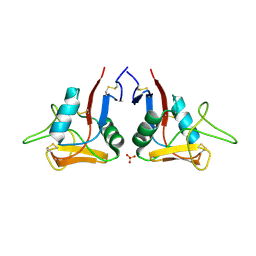 | | HUMAN CD69 - TETRAGONAL FORM | | Descriptor: | EARLY ACTIVATION ANTIGEN CD69, SULFATE ION | | Authors: | Tormo, J. | | Deposit date: | 2000-09-21 | | Release date: | 2000-09-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of the C-Type Lectin-Like Domain from the Human Hematopoietic Cell Receptor Cd69
J.Biol.Chem., 276, 2001
|
|
8W9J
 
 | | Crystal structure of human CLEC12A ectodomain complexed with 50C1 Fab | | Descriptor: | Anti-human CLEC12A antibody 50C1 heavy chain, Anti-human CLEC12A antibody 50C1 light chain, C-type lectin domain family 12 member A | | Authors: | Mori, S, Nagae, M, Yamasaki, S. | | Deposit date: | 2023-09-05 | | Release date: | 2024-03-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of the complex of CLEC12A and an antibody that interferes with binding of diverse ligands.
Int.Immunol., 36, 2024
|
|
