8SVI
 
 | | Ubiquitin variant i53:Mutant L67H with 53BP1 Tudor domain | | Descriptor: | GLYCEROL, Tumor protein p53 binding protein 1, Ubiquitin Variant i53: Mutant L67H | | Authors: | Partridge, J.R, Holden, J.K, Wibowo, A.S, Mulichak, A. | | Deposit date: | 2023-05-16 | | Release date: | 2024-03-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Functional screening in human HSPCs identifies optimized protein-based enhancers of Homology Directed Repair.
Nat Commun, 15, 2024
|
|
8SVH
 
 | | Ubiquitin variant i53 mutant L67R bound to 53BP1 Tudor Domain | | Descriptor: | Tumor protein p53 binding protein 1, Ubiquitin variant i53: mutant L67R | | Authors: | Holden, J.K, Partridge, J.R, Wibowo, A.S, Mulichak, A. | | Deposit date: | 2023-05-16 | | Release date: | 2024-03-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Functional screening in human HSPCs identifies optimized protein-based enhancers of Homology Directed Repair.
Nat Commun, 15, 2024
|
|
8SVG
 
 | | Ubiquitin variant i53 in complex with 53BP1 Tudor domain | | Descriptor: | Tumor protein p53 binding protein 1, Ubiquitin variant i53 | | Authors: | Holden, J.K, Partridge, J.R, Wibowo, A.S, Mulichak, A. | | Deposit date: | 2023-05-16 | | Release date: | 2024-03-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Functional screening in human HSPCs identifies optimized protein-based enhancers of Homology Directed Repair.
Nat Commun, 15, 2024
|
|
2G3R
 
 | | Crystal Structure of 53BP1 tandem tudor domains at 1.2 A resolution | | Descriptor: | SULFATE ION, Tumor suppressor p53-binding protein 1 | | Authors: | Lee, J, Botuyan, M.V, Thompson, J.R, Mer, G. | | Deposit date: | 2006-02-20 | | Release date: | 2007-01-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural Basis for the Methylation State-Specific Recognition of Histone H4-K20 by 53BP1 and Crb2 in DNA Repair.
Cell(Cambridge,Mass.), 127, 2006
|
|
6VA5
 
 | | Tudor Domain of Tumor suppressor p53BP1 with MFP-4184 | | Descriptor: | 2-(4-methylpiperazin-1-yl)aniline, GLYCEROL, SULFATE ION, ... | | Authors: | Zeng, H, Dong, A, Headey, S, Gunzburg, M, Doak, B, James, L.I, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-12-16 | | Release date: | 2020-04-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Tudor Domain of Tumor suppressor p53BP1 with MFP-4184
to be published
|
|
6VIP
 
 | | TUDOR DOMAIN OF TUMOR SUPPRESSOR P53BP1 WITH MFP-6008 | | Descriptor: | TP53-binding protein 1, UNKNOWN ATOM OR ION, {4-[(3,5-dimethyl-1H-pyrazol-1-yl)methyl]phenyl}(4-ethylpiperazin-1-yl)methanone | | Authors: | The, J, Hong, Z, Dong, A, Headey, S, Gunzburg, M, Doak, B, James, L.I, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-01-13 | | Release date: | 2020-02-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | TUDOR DOMAIN OF TUMOR SUPPRESSOR P53BP1 WITH MFP-6008
to be published
|
|
7LIN
 
 | |
4RG2
 
 | | Tudor Domain of Tumor suppressor p53BP1 with small molecule ligand | | Descriptor: | 1,2-ETHANEDIOL, 3-bromo-N-[3-(tert-butylamino)propyl]benzamide, Tumor suppressor p53-binding protein 1, ... | | Authors: | Dong, A, Mader, P, James, L, Perfetti, M, Tempel, W, Frye, S, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-09-29 | | Release date: | 2014-10-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Identification of a fragment-like small molecule ligand for the methyl-lysine binding protein, 53BP1.
ACS Chem. Biol., 10, 2015
|
|
3LGF
 
 | |
8SVJ
 
 | | Ubiquitin variant i53: mutant VHH with 53BP1 Tudor domain | | Descriptor: | GLYCEROL, Tumor protein p53 binding protein 1, Ubiquitin varient i53 mutant VHH | | Authors: | Holden, J, Partridge, J.R, Wibowo, A.S, Mulichak, A. | | Deposit date: | 2023-05-16 | | Release date: | 2024-03-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Functional screening in human HSPCs identifies optimized protein-based enhancers of Homology Directed Repair.
Nat Commun, 15, 2024
|
|
8F0W
 
 | | Tudor Domain of Tumor suppressor p53BP1 with MFP-5956 | | Descriptor: | 1-[4-(4-ethylpiperazin-1-yl)-3-fluorophenyl]butan-1-one, TP53-binding protein 1, UNKNOWN ATOM OR ION | | Authors: | The, J, Hong, Z, Dong, A, Headey, S, Gunzburg, M, Doak, B, James, L.I, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-11-04 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Tudor Domain of Tumor suppressor p53BP1 with MFP-5956
to be published
|
|
3LGL
 
 | |
8SWJ
 
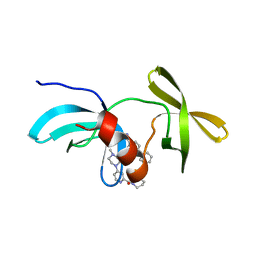 | | Co-crystal structure of 53BP1 tandem Tudor domains in complex with UNC8531 | | Descriptor: | (3S)-N-(4'-carbamoyl[1,1'-biphenyl]-3-yl)-1-[4-(4-methylpiperazin-1-yl)pyridine-2-carbonyl]piperidine-3-carboxamide, TP53-binding protein 1, UNKNOWN ATOM OR ION | | Authors: | Zeng, H, Dong, A, Shell, D.J, Foley, C, Pearce, K.H, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-05-18 | | Release date: | 2023-08-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Co-crystal structure of 53BP1 tandem Tudor domains in complex with UNC8531
To be published
|
|
6MXY
 
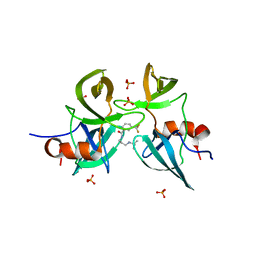 | |
6IUA
 
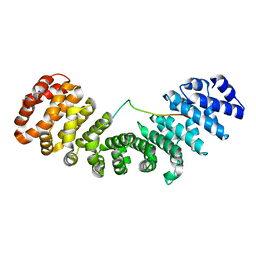 | |
8EOM
 
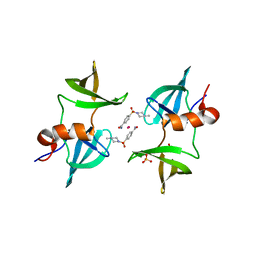 | | TUDOR DOMAIN OF TUMOR SUPPRESSOR P53BP1 WITH MFP-5973 | | Descriptor: | 4-(4-methylpiperazine-1-sulfonyl)benzamide, SULFATE ION, TP53-binding protein 1, ... | | Authors: | The, J, Hong, Z, Headey, S, Gunzburg, M, Doak, B, James, L.I, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-10-03 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | TUDOR DOMAIN OF TUMOR SUPPRESSOR P53BP1 WITH MFP-5973
to be published
|
|
2IG0
 
 | | Structure of 53BP1/methylated histone peptide complex | | Descriptor: | Dimethylated Histone H4-K20 peptide, SULFATE ION, Tumor suppressor p53-binding protein 1 | | Authors: | Mer, G. | | Deposit date: | 2006-09-22 | | Release date: | 2007-01-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis for the Methylation State-Specific Recognition of Histone H4-K20 by 53BP1 and Crb2 in DNA Repair.
Cell(Cambridge,Mass.), 127, 2006
|
|
8T2D
 
 | | Ubiquitin variant i53:Mutant T12Y.T14E.L67R with 53BP1 Tudor domain | | Descriptor: | Tumor protein p53 binding protein 1, Ubiquitin variant i53 | | Authors: | Partridge, J.R, Holden, J.K, Wibowo, A.S, Mulichak, A. | | Deposit date: | 2023-06-05 | | Release date: | 2024-03-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.751 Å) | | Cite: | Functional screening in human HSPCs identifies optimized protein-based enhancers of Homology Directed Repair.
Nat Commun, 15, 2024
|
|
5Z78
 
 | | Structure of TIRR/53BP1 complex | | Descriptor: | TP53-binding protein 1, Tudor-interacting repair regulator protein | | Authors: | Dai, Y.X, Shan, S. | | Deposit date: | 2018-01-27 | | Release date: | 2018-06-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.762 Å) | | Cite: | Structural basis for recognition of 53BP1 tandem Tudor domain by TIRR
Nat Commun, 9, 2018
|
|
4X34
 
 | |
8HKW
 
 | |
3LH0
 
 | |
6IU7
 
 | |
6UPT
 
 | | Tudor Domain of Tumor suppressor p53BP1 with MFP-2706 | | Descriptor: | 2-((2-chlorobenzyl)thio)-4,5-dihydro-1H-imidazole, TP53-binding protein 1, UNKNOWN ATOM OR ION | | Authors: | The, J, Dong, A, Headey, S, Gunzburg, M, Doak, B, James, L.I, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-10-18 | | Release date: | 2019-11-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Tudor Domain of Tumor suppressor p53BP1 with MFP-2706
to be published
|
|
5ZCJ
 
 | | Crystal structure of complex | | Descriptor: | TP53-binding protein 1, Tudor-interacting repair regulator protein | | Authors: | Wang, J, Yuan, Z, Cui, Y, Xie, R, Wang, M, Ma, Y, Yu, X, Liu, X. | | Deposit date: | 2018-02-17 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.004 Å) | | Cite: | Crystal structure of complex
To Be Published
|
|
