7DE9
 
 | |
5YVX
 
 | | Crystal structure of SDG8 CW domain in complex with H3K4me1 peptide | | Descriptor: | H3K4me1, Histone-lysine N-methyltransferase ASHH2, ZINC ION | | Authors: | Liu, Y, Huang, Y. | | Deposit date: | 2017-11-27 | | Release date: | 2018-03-14 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.591 Å) | | Cite: | Uncovering the mechanistic basis for specific recognition of monomethylated H3K4 by the CW domain ofArabidopsishistone methyltransferase SDG8.
J. Biol. Chem., 293, 2018
|
|
5B73
 
 | | Crystal structure of human ZMYND8 PHD-Bromo-PWWP domain | | Descriptor: | Protein kinase C-binding protein 1, ZINC ION | | Authors: | Li, H, Li, Y, Zheng, X. | | Deposit date: | 2016-06-03 | | Release date: | 2016-11-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | ZMYND8 Reads the Dual Histone Mark H3K4me1-H3K14ac to Antagonize the Expression of Metastasis-Linked Genes
Mol.Cell, 63, 2016
|
|
5SZC
 
 | | Structure of human Dpf3 double-PHD domain bound to histone H3 tail peptide with monomethylated K4 and acetylated K14 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Histone H3 tail peptide, ZINC ION, ... | | Authors: | Singh, N, Local, A, Shiau, A, Ren, B, Corbett, K.D. | | Deposit date: | 2016-08-13 | | Release date: | 2017-08-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.193 Å) | | Cite: | Identification of H3K4me1-associated proteins at mammalian enhancers.
Nat. Genet., 50, 2018
|
|
5SZB
 
 | | Structure of human Dpf3 double-PHD domain bound to histone H3 tail peptide with acetylated K14 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Histone H3 tail peptide, ZINC ION, ... | | Authors: | Singh, N, Local, A, Shiau, A, Ren, B, Corbett, K.D. | | Deposit date: | 2016-08-13 | | Release date: | 2017-08-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Identification of H3K4me1-associated proteins at mammalian enhancers.
Nat. Genet., 50, 2018
|
|
5SVY
 
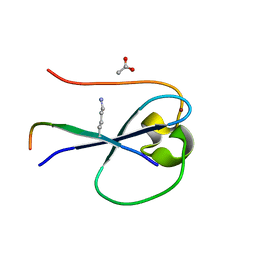 | | MORC3 CW in complex with histone H3K4me1 | | Descriptor: | ACETATE ION, H3K4me1, MORC family CW-type zinc finger protein 3, ... | | Authors: | Tong, Q, Andrews, F.H, Kutateladze, T.G. | | Deposit date: | 2016-08-08 | | Release date: | 2016-10-05 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Multivalent Chromatin Engagement and Inter-domain Crosstalk Regulate MORC3 ATPase.
Cell Rep, 16, 2016
|
|
6QXZ
 
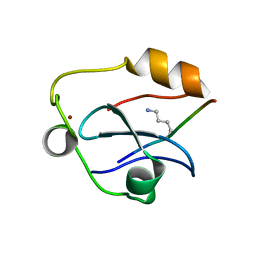 | | Solution structure of the ASHH2 CW domain with the N-terminal histone H3 tail mimicking peptide monomethylated on lysine 4 | | Descriptor: | ALA-ARG-THR-MLZ-GLN-THR-ALA-ARG-TYR, Histone-lysine N-methyltransferase ASHH2, ZINC ION | | Authors: | Dobrovolska, O, Madeleine, N, Teigen, K, Halskau, O, Bril'kov, M. | | Deposit date: | 2019-03-08 | | Release date: | 2019-12-04 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The Arabidopsis (ASHH2) CW domain binds monomethylated K4 of the histone H3 tail through conformational selection.
Febs J., 287, 2020
|
|
7W6I
 
 | | The crystal structure of MLL1 (N3861I/Q3867L/C3882SS)-RBBP5-ASH2L in complex with H3K4me1 peptide | | Descriptor: | Histone H3.3C, Histone-lysine N-methyltransferase 2A, Retinoblastoma-binding protein 5, ... | | Authors: | Zhao, L, Li, Y, Chen, Y. | | Deposit date: | 2021-12-01 | | Release date: | 2022-09-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Structural basis for product specificities of MLL family methyltransferases.
Mol.Cell, 82, 2022
|
|
4GUU
 
 | | Crystal structure of LSD2-NPAC with tranylcypromine | | Descriptor: | Lysine-specific histone demethylase 1B, Putative oxidoreductase GLYR1, ZINC ION, ... | | Authors: | Chen, F, Dong, Z, Fang, J, Yang, Y, Li, Z, Xu, Y, Yang, H, Wang, P, Xu, Y. | | Deposit date: | 2012-08-29 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | LSD2/KDM1B and its cofactor NPAC/GLYR1 endow a structural and molecular model for regulation of H3K4 demethylation
Mol.Cell, 49, 2013
|
|
4GUS
 
 | | Crystal structure of LSD2-NPAC with H3 in space group P3221 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Histone H3.3, ... | | Authors: | Chen, F, Dong, Z, Fang, J, Yang, Y, Li, Z, Xu, Y, Yang, H, Wang, P, Fang, R, Shi, Y, Xu, Y. | | Deposit date: | 2012-08-29 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | LSD2/KDM1B and its cofactor NPAC/GLYR1 endow a structural and molecular model for regulation of H3K4 demethylation
Mol.Cell, 49, 2013
|
|
4GUR
 
 | | Crystal structure of LSD2-NPAC with H3 in space group P21 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Histone H3.3, ... | | Authors: | Chen, F, Dong, Z, Fang, J, Yang, Y, Li, Z, Xu, Y, Yang, H, Wang, P, Fang, R, Shi, Y, Xu, Y. | | Deposit date: | 2012-08-29 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.506 Å) | | Cite: | LSD2/KDM1B and its cofactor NPAC/GLYR1 endow a structural and molecular model for regulation of H3K4 demethylation
Mol.Cell, 49, 2013
|
|
4GUT
 
 | | Crystal structure of LSD2-NPAC | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Lysine-specific histone demethylase 1B, ... | | Authors: | Chen, F, Dong, Z, Fang, J, Yang, Y, Li, Z, Xu, Y, Yang, H, Wang, P, Fang, R, Shi, Y, Xu, Y. | | Deposit date: | 2012-08-29 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | LSD2/KDM1B and its cofactor NPAC/GLYR1 endow a structural and molecular model for regulation of H3K4 demethylation
Mol.Cell, 49, 2013
|
|
4IUV
 
 | |
8HMX
 
 | |
8Q1N
 
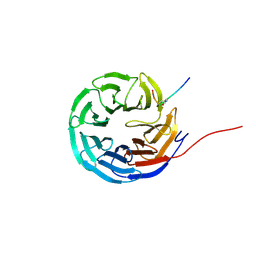 | | Cyclic peptide binder of the WBM-site of WDR5 | | Descriptor: | Cyclic peptide inhibitor, WD repeat-containing protein 5 | | Authors: | Schmeing, S, Chang, J.Y, t Hart, P, Gasper, R. | | Deposit date: | 2023-08-01 | | Release date: | 2023-09-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.843 Å) | | Cite: | Macrocyclic peptides as inhibitors of WDR5-lncRNA interactions.
Chem.Commun.(Camb.), 59, 2023
|
|
7Q2J
 
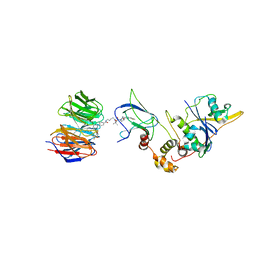 | | Quaternary Complex of human WDR5 and pVHL:ElonginC:ElonginB bound to PROTAC Homer | | Descriptor: | Elongin-B, Elongin-C, N-[5-[4-[[5-[[(2S)-3,3-dimethyl-1-[(2S,4R)-2-[[4-(4-methyl-1,3-thiazol-5-yl)phenyl]methylcarbamoyl]-4-oxidanyl-pyrrolidin-1-yl]-1-oxidanylidene-butan-2-yl]amino]-5-oxidanylidene-pentyl]carbamoyl]phenyl]-2-(4-methylpiperazin-1-yl)phenyl]-6-oxidanylidene-4-(trifluoromethyl)-1H-pyridine-3-carboxamide, ... | | Authors: | Kraemer, A, Doelle, A, Schwalm, M.P, Adhikari, B, Wolf, E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-10-25 | | Release date: | 2021-11-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Tracking the PROTAC degradation pathway in living cells highlights the importance of ternary complex measurement for PROTAC optimization.
Cell Chem Biol, 30, 2023
|
|
4A7J
 
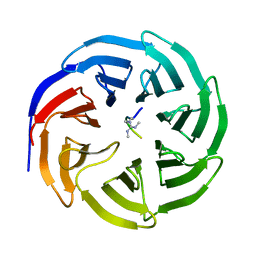 | | Symmetric Dimethylation of H3 Arginine 2 is a Novel Histone Mark that Supports Euchromatin Maintenance | | Descriptor: | HISTONE H3.1T, WD REPEAT-CONTAINING PROTEIN 5 | | Authors: | Migliori, V, Muller, J, Phalke, S, Low, D, Bezzi, M, ChuenMok, W, Gunaratne, J, Capasso, P, Bassi, C, Cecatiello, V, DeMarco, A, Blackstock, W, Kuznetsov, V, Amati, B, Mapelli, M, Guccione, E. | | Deposit date: | 2011-11-14 | | Release date: | 2012-01-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Symmetric Dimethylation of H3R2 is a Newly Identified Histone Mark that Supports Euchromatin Maintenance
Nat.Struct.Mol.Biol., 19, 2012
|
|
4Y7R
 
 | | Crystal structure of WDR5 in complex with MYC MbIIIb peptide | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MYC MbIIIb peptide, ... | | Authors: | Sun, Q, Phan, J, Olejniczak, E.T, Thomas, L.R, Fesik, S.W, Tansey, W.P. | | Deposit date: | 2015-02-16 | | Release date: | 2015-04-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Interaction with WDR5 Promotes Target Gene Recognition and Tumorigenesis by MYC.
Mol.Cell, 58, 2015
|
|
4O45
 
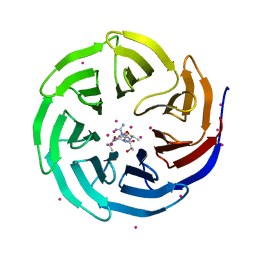 | | WDR5 in complex with influenza NS1 C-terminal tail | | Descriptor: | Nonstructural protein 1, UNKNOWN ATOM OR ION, WD repeat-containing protein 5 | | Authors: | Qin, S, Xu, C, Tempel, W, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-12-18 | | Release date: | 2014-04-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural basis for histone mimicry and hijacking of host proteins by influenza virus protein NS1.
Nat Commun, 5, 2014
|
|
5VFC
 
 | | WDR5 bound to inhibitor MM-589 | | Descriptor: | 1,2-ETHANEDIOL, N-{(3R,6S,9S,12R)-6-ethyl-12-methyl-9-[3-(N'-methylcarbamimidamido)propyl]-2,5,8,11-tetraoxo-3-phenyl-1,4,7,10-tetraazacyclotetradecan-12-yl}-2-methylpropanamide, SULFATE ION, ... | | Authors: | Stuckey, J.A. | | Deposit date: | 2017-04-07 | | Release date: | 2017-06-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Discovery of a Highly Potent, Cell-Permeable Macrocyclic Peptidomimetic (MM-589) Targeting the WD Repeat Domain 5 Protein (WDR5)-Mixed Lineage Leukemia (MLL) Protein-Protein Interaction.
J. Med. Chem., 60, 2017
|
|
6WJQ
 
 | |
2O9K
 
 | | WDR5 in Complex with Dimethylated H3K4 Peptide | | Descriptor: | H3 HISTONE, WD repeat protein 5 | | Authors: | Min, J.R, Schuetz, A, Allali-Hassani, A, Martin, F, Loppnau, P, Vedadi, M, Weigelt, J, Sundstrom, M, Edwards, A.M, Arrowsmith, C.H, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-12-13 | | Release date: | 2006-12-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for Molecular Recognition and Presentation of Histone H3 by Wdr5.
Embo J., 25, 2006
|
|
6OI0
 
 | | Crystal structure of human WDR5 in complex with L-arginine | | Descriptor: | ARGININE, GLYCEROL, SULFATE ION, ... | | Authors: | Lorton, B.M, Harijan, R.K, Burgos, E, Bonanno, J.B, Almo, S.C, Shechter, D. | | Deposit date: | 2019-04-08 | | Release date: | 2020-04-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | A Binary Arginine Methylation Switch on Histone H3 Arginine 2 Regulates Its Interaction with WDR5.
Biochemistry, 59, 2020
|
|
4CY1
 
 | | Crystal structure of the KANSL1-WDR5 complex. | | Descriptor: | GLYCEROL, KAT8 REGULATORY NSL COMPLEX SUBUNIT 1, WD REPEAT-CONTAINING PROTEIN 5 | | Authors: | Dias, J, Brettschneider, J, Cusack, S, Kadlec, J. | | Deposit date: | 2014-04-09 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Analysis of the Kansl1/Wdr5/Kansl2 Complex Reveals that Wdr5 is Required for Efficient Assembly and Chromatin Targeting of the Nsl Complex.
Genes Dev., 28, 2014
|
|
4CY2
 
 | | Crystal structure of the KANSL1-WDR5-KANSL2 complex. | | Descriptor: | KAT8 REGULATORY NSL COMPLEX SUBUNIT 1, KAT8 REGULATORY NSL COMPLEX SUBUNIT 2, WD REPEAT-CONTAINING PROTEIN 5 | | Authors: | Dias, J, Brettschneider, J, Cusack, S, Kadlec, J. | | Deposit date: | 2014-04-09 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Analysis of the Kansl1/Wdr5/Kansl2 Complex Reveals that Wdr5 is Required for Efficient Assembly and Chromatin Targeting of the Nsl Complex.
Genes Dev., 28, 2014
|
|
