6TW5
 
 | | Plasmodium vivax N-myristoyltransferase with bound indazole inhibitor IMP-917 | | Descriptor: | 1-[5-[4-fluoranyl-2-[2-(1,3,5-trimethylpyrazol-4-yl)ethoxy]phenyl]-2~{H}-indazol-3-yl]-~{N},~{N}-dimethyl-methanamine, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Brannigan, J.A. | | Deposit date: | 2020-01-12 | | Release date: | 2021-07-21 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Plasmodium vivax N-myristoyltransferase with bound indazole inhibitor IMP-917
To Be Published
|
|
7VJK
 
 | | class II photolyase MmCPDII semiquinone to fully reduced TR-SFX studies (100 us time-point) | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, DNA photolyase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Maestre-Reyna, M, Yang, C.-H, Huang, W.-C, Nango, E, Ngura Putu, E.P.G, Franz-Badur, S, Wu, W.-J, Wu, H.-Y, Wang, P.-H, Hosokawa, Y, Saft, M, Emmerich, H.-J, Liao, J.-H, Lee, C.-C, Huang, K.-F, Chang, Y.-K, Weng, J.-H, Royant, A, Gad, W, Pang, A.H, Chang, C.-W, Sugahara, M, Owada, S, Joti, Y, Yamashita, A, Tanaka, R, Tanaka, T, Luo, F.J, Tono, K, Kiontke, S, Yamamoto, J, Iwata, S, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2021-09-28 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Serial crystallography captures dynamic control of sequential electron and proton transfer events in a flavoenzyme.
Nat.Chem., 14, 2022
|
|
7TA9
 
 | |
5B8H
 
 | |
7W44
 
 | | Complex structure of a leaf-branch compost cutinase variant LCC ICCG_RIP | | Descriptor: | 1,2-ETHANEDIOL, IMIDAZOLE, Leaf-branch compost cutinase | | Authors: | Niu, D, Zeng, W, Huang, J.W, Chen, C.C, Liu, W.D, Guo, R.T. | | Deposit date: | 2021-11-26 | | Release date: | 2022-03-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Substrate-Binding Mode of a Thermophilic PET Hydrolase and Engineering the Enzyme to Enhance the Hydrolytic Efficacy.
Acs Catalysis, 12, 2022
|
|
7ONS
 
 | | PARP1 catalytic domain in complex with isoquinolone-based inhibitor (compound 16) | | Descriptor: | 7-[[4-(1,5-dimethylimidazol-2-yl)piperazin-1-yl]methyl]-3-ethyl-1~{H}-quinolin-2-one, Poly [ADP-ribose] polymerase 1, SULFATE ION | | Authors: | Schimpl, M, Balazs, A, Barratt, D, Bista, M, Chuba, M, Degorce, S.L, Di Fruscia, P, Embrey, K, Ghosh, A, Gill, S, Gunnarsson, A, Hande, S, Hemsley, P, Heightman, T.D, Illuzzi, G, Lane, J, Larner, C, Leo, E, Madin, A, Martin, S, McWilliams, L, Orme, J, Pachl, F, Packer, M, Pike, A, Staniszewska, A.D, Talbot, V, Underwood, E, Varnes, G.J, Zhang, A, Zheng, X, Johannes, J.W. | | Deposit date: | 2021-05-25 | | Release date: | 2021-09-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Discovery of 5-{4-[(7-Ethyl-6-oxo-5,6-dihydro-1,5-naphthyridin-3-yl)methyl]piperazin-1-yl}- N -methylpyridine-2-carboxamide (AZD5305): A PARP1-DNA Trapper with High Selectivity for PARP1 over PARP2 and Other PARPs.
J.Med.Chem., 64, 2021
|
|
8B4U
 
 | | The crystal structure of PET46, a PETase enzyme from Candidatus bathyarchaeota | | Descriptor: | 1,2-ETHANEDIOL, Alpha/beta hydrolase, CHLORIDE ION, ... | | Authors: | Costanzi, E, Applegate, V, Schumacher, J, Smits, S.H.J. | | Deposit date: | 2022-09-21 | | Release date: | 2023-08-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | An archaeal lid-containing feruloyl esterase degrades polyethylene terephthalate.
Commun Chem, 6, 2023
|
|
7VJ7
 
 | | SFX structure of archaeal class II CPD photolyase from Methanosarcina mazei in the fully reduced state | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, DNA photolyase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Maestre-Reyna, M, Yang, C.-H, Huang, W.C, Nango, E, Gusti-Ngurah-Putu, E.-P, Franz-Badur, S, Wu, W.-J, Wu, H.-Y, Wang, P.-H, Liao, J.-H, Lee, C.-C, Huang, K.-F, Chang, Y.-K, Weng, J.-H, Sugahara, M, Owada, S, Joti, Y, Tanaka, R, Tono, K, Kiontke, S, Yamamoto, J, Iwata, S, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2021-09-28 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Serial crystallography captures dynamic control of sequential electron and proton transfer events in a flavoenzyme.
Nat.Chem., 14, 2022
|
|
7VJJ
 
 | | class II photolyase MmCPDII semiquinone to fully reduced TR-SFX studies (30 us time-point) | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, DNA photolyase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Maestre-Reyna, M, Yang, C.-H, Huang, W.-C, Nango, E, Ngura Putu, E.P.G, Franz-Badur, S, Wu, W.-J, Wu, H.-Y, Wang, P.-H, Hosokawa, Y, Saft, M, Emmerich, H.-J, Liao, J.-H, Lee, C.-C, Huang, K.-F, Chang, Y.-K, Weng, J.-H, Royant, A, Gad, W, Pang, A.H, Chang, C.-W, Sugahara, M, Owada, S, Joti, Y, Yamashita, A, Tanaka, R, Tanaka, T, Luo, F.J, Tono, K, Kiontke, S, Yamamoto, J, Iwata, S, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2021-09-28 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Serial crystallography captures dynamic control of sequential electron and proton transfer events in a flavoenzyme.
Nat.Chem., 14, 2022
|
|
8RMY
 
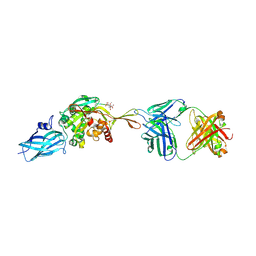 | |
8AQF
 
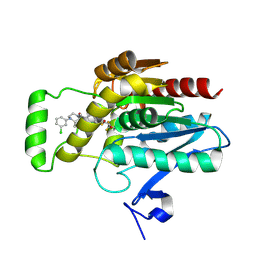 | | CRYSTAL STRUCTURE OF HUMAN MONOGLYCERIDE LIPASE WITH COMPOUND LEI-515 | | Descriptor: | 1-[(~{R})-[2-chloranyl-4-[(2~{S},3~{S})-4-(3-chlorophenyl)-2,3-dimethyl-piperazin-1-yl]carbonyl-phenyl]sulfinyl]-3,3-bis(fluoranyl)pentan-2-one, Monoglyceride lipase | | Authors: | Jiang, M, Huizenga, M, Wirt, J, Paloczi, J, Amedi, A, van der Berg, R, Benz, J, Collin, L, Deng, H, Driever, W, Florea, B, Grether, U, Janssen, A, Heitman, L, Lam, T.W, Mohr, F, Pavlovic, A, Ruf, I, Rutjes, H, Stevens, F, van der Vliet, D, van der Wel, T, Wittwer, M, Boeckel, C, Pacher, P, Hohmann, A, van der Stelt, M. | | Deposit date: | 2022-08-12 | | Release date: | 2023-08-23 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A monoacylglycerol lipase inhibitor showing therapeutic efficacy in mice without central side effects or dependence.
Nat Commun, 14, 2023
|
|
7ONT
 
 | | PARP1 catalytic domain in complex with a selective pyridine carboxamide-based inhibitor (compound 22) | | Descriptor: | 5-[4-[(3-ethyl-2-oxidanylidene-1~{H}-quinolin-7-yl)methyl]piperazin-1-yl]-~{N}-methyl-pyridine-2-carboxamide, Poly [ADP-ribose] polymerase 1, SULFATE ION | | Authors: | Schimpl, M, Balazs, A, Barratt, D, Bista, M, Chuba, M, Degorce, S.L, Di Fruscia, P, Embrey, K, Ghosh, A, Gill, S, Gunnarsson, A, Hande, S, Hemsley, P, Heightman, T.D, Illuzzi, G, Lane, J, Larner, C, Leo, E, Madin, A, Martin, S, McWilliams, L, Orme, J, Pachl, F, Packer, M.J, Pike, A, Staniszewska, A.D, Talbot, V, Underwood, E, Varnes, G.J, Zhang, A, Zheng, X, Johannes, J.W. | | Deposit date: | 2021-05-25 | | Release date: | 2021-09-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.853 Å) | | Cite: | Discovery of 5-{4-[(7-Ethyl-6-oxo-5,6-dihydro-1,5-naphthyridin-3-yl)methyl]piperazin-1-yl}- N -methylpyridine-2-carboxamide (AZD5305): A PARP1-DNA Trapper with High Selectivity for PARP1 over PARP2 and Other PARPs.
J.Med.Chem., 64, 2021
|
|
6HXF
 
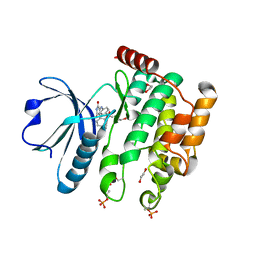 | | Human STK10 bound to a maleimide inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 3-(2-methoxyphenyl)-4-[[4-(phenylcarbonyl)phenyl]amino]pyrrole-2,5-dione, CHLORIDE ION, ... | | Authors: | Sorrell, F.J, Salah, E, Serafim, R.A.M, Savitsky, P.A, Krojer, T, Bailey, H.J, Pinkas, D, Burgess-Brown, N.A, von Delft, F, Knapp, S, Arrowsmith, C, Bountra, C, Edwards, A.M, Elkins, J.M. | | Deposit date: | 2018-10-17 | | Release date: | 2018-10-31 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Human STK10 bound to a maleimide inhibitor
To Be Published
|
|
5ZW9
 
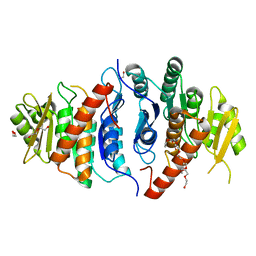 | | Crystal structure of Pyridoxal kinase (PdxK) from Salmonella typhimurium | | Descriptor: | 1,2-ETHANEDIOL, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Deka, G, Benazir, J.F, Kalyani, J.N, Savithri, H.S, Murthy, M.R.N. | | Deposit date: | 2018-05-14 | | Release date: | 2019-05-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and functional studies on Salmonella typhimurium pyridoxal kinase: the first structural evidence for the formation of Schiff base with the substrate.
Febs J., 286, 2019
|
|
8ANB
 
 | |
9FWZ
 
 | | Cryo-EM structure of the type 1 pilus assembly platform as part of the FimA-bound chaperone-usher pilus complex (Local refinement including FimD, FimC, FimAn, FimAn-1 and FimAn-2) | | Descriptor: | Chaperone protein FimC, Outer membrane usher protein FimD, Type-1 fimbrial protein, ... | | Authors: | Bachmann, P, Afanasyev, P, Boehringer, D, Glockshuber, R. | | Deposit date: | 2024-07-01 | | Release date: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structure of the type 1 pilus assembly platform as part of the FimA-bound chaperone-usher pilus complex (Local refinement including FimD, FimC, FimAn, FimAn-1 and FimAn-2)
To Be Published
|
|
5LRD
 
 | | Crystal structure of Glycogen Phosphorylase b in complex with KS242 | | Descriptor: | (2~{R},3~{S},4~{R},5~{R},6~{S})-2-(hydroxymethyl)-6-[5-(4-methylphenyl)-4~{H}-1,2,4-triazol-3-yl]oxane-3,4,5-triol, DIMETHYL SULFOXIDE, Glycogen phosphorylase, ... | | Authors: | Kantsadi, A.L, Leonidas, D.D. | | Deposit date: | 2016-08-18 | | Release date: | 2017-06-14 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | van der Waals interactions govern C-beta-d-glucopyranosyl triazoles' nM inhibitory potency in human liver glycogen phosphorylase.
J. Struct. Biol., 199, 2017
|
|
6H8S
 
 | |
5KLB
 
 | | Crystal structure of the CavAb voltage-gated calcium channel(wild-type, 2.7A) | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, CALCIUM ION, ... | | Authors: | Tang, L, Gamal EL-Din, T.M, Swanson, T.M, Pryde, D.C, Scheuer, T, Zheng, N, Catterall, W.A. | | Deposit date: | 2016-06-23 | | Release date: | 2016-08-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for inhibition of a voltage-gated Ca(2+) channel by Ca(2+) antagonist drugs.
Nature, 537, 2016
|
|
9FWB
 
 | | Cryo-EM structure of the type 1 pilus assembly platform as part of the FimA-bound chaperone-usher pilus complex (Local refinement including FimD, FimC, FimAn and FimAn-1) | | Descriptor: | Chaperone protein FimC, Outer membrane usher protein FimD, Type-1 fimbrial protein, ... | | Authors: | Bachmann, P, Afanasyev, P, Boehringer, D, Glockshuber, R. | | Deposit date: | 2024-06-28 | | Release date: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structure of the type 1 pilus assembly platform as part of the FimA-bound chaperone-usher pilus complex (Local refinement including FimD, FimC, FimAn and FimAn-1)
To Be Published
|
|
9FY9
 
 | | Cryo-EM structure of the type 1 chaperone-usher pilus FimD-tip complex (FimDHGFC) - Conformer 1 | | Descriptor: | Chaperone protein FimC, Outer membrane usher protein FimD, Protein FimF, ... | | Authors: | Bachmann, P, Afanasyev, P, Boehringer, D, Glockshuber, R. | | Deposit date: | 2024-07-03 | | Release date: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of the type 1 chaperone-usher pilus FimD-tip complex (FimDHGFC) - Conformer 1
To Be Published
|
|
5KLS
 
 | | Structure of CavAb in complex with Br-dihydropyridine derivative UK-59811 | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, CALCIUM ION, Ion transport protein, ... | | Authors: | Tang, L, Gamal EL-Din, T.M, Swanson, T.M, Pryde, D.C, Scheuer, T, Zheng, N, Catterall, W.A. | | Deposit date: | 2016-06-25 | | Release date: | 2016-08-31 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.299 Å) | | Cite: | Structural basis for inhibition of a voltage-gated Ca(2+) channel by Ca(2+) antagonist drugs.
Nature, 537, 2016
|
|
8SK2
 
 | |
5BSZ
 
 | | X-ray structure of the sugar N-methyltransferase KedS8 from Streptoalloteichus sp ATCC 53650 | | Descriptor: | 1,2-ETHANEDIOL, N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Thoden, J.B, Delvaux, N.A, Holden, H.M. | | Deposit date: | 2015-06-02 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular architecture of KedS8, a sugar N-methyltransferase from Streptoalloteichus sp. ATCC 53650.
Protein Sci., 24, 2015
|
|
9JML
 
 | | NADP-dependent oxidoreductase complexed with NADP and substrate 1 | | Descriptor: | (1Z,3Z,5Z,7S,8R,9S,10S,11R,13R,15R,16Z,18Z,24S)-11-ethyl-2,7-dihydroxy-10-methyl-21,25-diazatetracyclo[22.2.1.08,15.09,13]heptacosa-1,3,5,16,18-pentaene-20,26,27-trione, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADP-dependent oxidoreductase | | Authors: | Li, Y, Zhu, D, Xie, X, Li, F, Lu, M. | | Deposit date: | 2024-09-20 | | Release date: | 2025-06-18 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structural Basis for Medium-Chain Dehydrogenase/Reductase-Catalyzed Reductive Cyclization in Polycyclic Tetramate Macrolactam Biosynthesis.
J.Am.Chem.Soc., 147, 2025
|
|
