8T0S
 
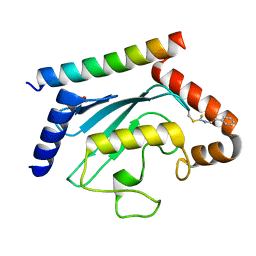 | | Crystal structure of UBE2G2 adduct with phenethyl isothiocyanate (PEITC) at the Cys48 position | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase AMFR, Ubiquitin-conjugating enzyme E2 G2 | | Authors: | Wang, C, Shaw, G.X, Shi, G, Ji, X. | | Deposit date: | 2023-06-01 | | Release date: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of UBE2G2 adduct with phenethyl isothiocyanate (PEITC) at the Cys48 position
To be published
|
|
2ZKN
 
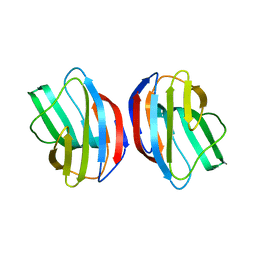 | |
6ATE
 
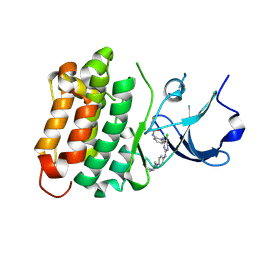 | | SRC kinase bound to covalent inhibitor | | Descriptor: | N-{2-[(5-chloro-2-{[4-(4-methylpiperazin-1-yl)phenyl]amino}pyrimidin-4-yl)amino]phenyl}propanamide, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Gurbani, D, Westover, K.D. | | Deposit date: | 2017-08-28 | | Release date: | 2019-02-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.402 Å) | | Cite: | Leveraging Compound Promiscuity to Identify Targetable Cysteines within the Kinome.
Cell Chem Biol, 26, 2019
|
|
4G35
 
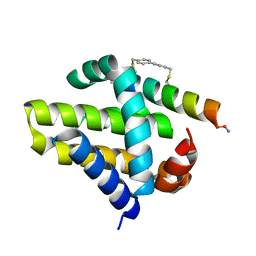 | | Mcl-1 in complex with a biphenyl cross-linked Noxa peptide. | | Descriptor: | 4,4'-bis(bromomethyl)biphenyl, Induced myeloid leukemia cell differentiation protein Mcl-1 homolog, Noxa BH3 peptide (cysteine-mediated cross-linked) | | Authors: | Drake, E, Edwardraja, S, Lin, Q, Gulick, A.M. | | Deposit date: | 2012-07-13 | | Release date: | 2012-12-05 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Rational design of proteolytically stable, cell-permeable peptide-based selective Mcl-1 inhibitors.
J.Am.Chem.Soc., 134, 2012
|
|
4NCT
 
 | | Human DYRK1A in complex with PKC412 | | Descriptor: | Dual specificity tyrosine-phosphorylation-regulated kinase 1A, PKC412 | | Authors: | Alexeeva, M.O, Rothweiler, U. | | Deposit date: | 2013-10-25 | | Release date: | 2015-04-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.597 Å) | | Cite: | The structure of a dual-specificity tyrosine phosphorylation-regulated kinase 1A-PKC412 complex reveals disulfide-bridge formation with the anomalous catalytic loop HRD(HCD) cysteine.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4A00
 
 | | Structure of an engineered aspartate aminotransferase | | Descriptor: | ALANYL-PYRIDOXAL-5'-PHOSPHATE, ASPARTATE AMINOTRANSFERASE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Fernandez, F.J, deVries, D, Pena-Soler, E, Coll, M, Christen, P, Gehring, H, Vega, M.C. | | Deposit date: | 2011-09-06 | | Release date: | 2011-12-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structure and Mechanism of a Cysteine Sulfinate Desulfinase Engineered on the Aspartate Aminotransferase Scaffold.
Biocim.Biophys.Acta, 1824, 2011
|
|
6XYI
 
 | | NMR solution structure of alpha-AnmTX- Ms11a-3 (Ms11a-3) | | Descriptor: | AMS9.3.1 | | Authors: | Mineev, K.S, Kornilov, F.D, Lushpa, V.A, Logashina, Y.A, Maleeva, E.E, Andreev, Y.A. | | Deposit date: | 2020-01-30 | | Release date: | 2021-02-10 | | Last modified: | 2025-02-19 | | Method: | SOLUTION NMR | | Cite: | Peptides from the Sea Anemone Metridium senile with Modified Inhibitor Cystine Knot (ICK) Fold Inhibit Nicotinic Acetylcholine Receptors.
Toxins, 15, 2022
|
|
3ZZK
 
 | | Structure of an engineered aspartate aminotransferase | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, ASPARTATE AMINOTRANSFERASE, GLYCEROL, ... | | Authors: | Fernandez, F.J, deVries, D, Pena-Soler, E, Coll, M, Christen, P, Gehring, H, Vega, M.C. | | Deposit date: | 2011-09-01 | | Release date: | 2011-12-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure and Mechanism of a Cysteine Sulfinate Desulfinase Engineered on the Aspartate Aminotransferase Scaffold.
Biocim.Biophys.Acta, 1824, 2011
|
|
4C8R
 
 | | Human gamma-butyrobetaine dioxygenase (BBOX1) in complex with Ni(II) and N-(3-hydroxypicolinoyl)-S-(pyridin-2-ylmethyl)-L-cysteine (AR692B) | | Descriptor: | 1,2-ETHANEDIOL, GAMMA-BUTYROBETAINE DIOXYGENASE, N-(3-hydroxypicolinoyl)-S-(pyridin-2-ylmethyl)-L-cysteine, ... | | Authors: | Chowdhury, R, Rydzik, A.M, Kochan, G.T, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2013-10-01 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Modulating carnitine levels by targeting its biosynthesis pathway - selective inhibition of gamma-butyrobetaine hydroxylase.
Chem Sci, 5, 2014
|
|
1KFP
 
 | | Solution structure of the antimicrobial 18-residue gomesin | | Descriptor: | GOMESIN | | Authors: | Mandard, N, Bulet, P, Caille, A, Daffre, S, Vovelle, F. | | Deposit date: | 2001-11-22 | | Release date: | 2002-04-10 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | The solution structure of gomesin, an antimicrobial cysteine-rich peptide from the spider.
Eur.J.Biochem., 269, 2002
|
|
4D9T
 
 | | Rsk2 C-terminal Kinase Domain with inhibitor (E)-methyl 3-(4-amino-7-(3-hydroxypropyl)-5-p-tolyl-7H-pyrrolo[2,3-d]pyrimidin-6-yl)-2-cyanoacrylate | | Descriptor: | Ribosomal protein S6 kinase alpha-3, SODIUM ION, methyl (2S)-3-{4-amino-7-[(1E)-3-hydroxyprop-1-en-1-yl]-5-(4-methylphenyl)-7H-pyrrolo[2,3-d]pyrimidin-6-yl}-2-cyanopropanoate | | Authors: | Serafimova, I.M, Pufall, M.A, Krishnan, S, Duda, K, Cohen, M.S, Maglathlin, R.L, McFarland, J.M, Miller, R.M, Frodin, M, Taunton, J. | | Deposit date: | 2012-01-12 | | Release date: | 2012-04-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Reversible targeting of noncatalytic cysteines with chemically tuned electrophiles.
Nat.Chem.Biol., 8, 2012
|
|
6WQ3
 
 | | Crystal Structure of Nsp16-Nsp10 Heterodimer from SARS-CoV-2 in Complex with 7-methyl-GpppA and S-adenosyl-L-homocysteine. | | Descriptor: | 2'-O-methyltransferase, 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, Non-structural protein 10, ... | | Authors: | Minasov, G, Shuvalova, L, Rosas-Lemus, M, Brunzelle, J.S, Kiryukhina, O, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-04-28 | | Release date: | 2020-05-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | High-resolution structures of the SARS-CoV-2 2'- O -methyltransferase reveal strategies for structure-based inhibitor design.
Sci.Signal., 13, 2020
|
|
7POY
 
 | | Spin labeled IPNS S55C variant in complex with Fe, ACV and NO | | Descriptor: | FE (III) ION, Isopenicillin N synthase, L-D-(A-AMINOADIPOYL)-L-CYSTEINYL-D-VALINE, ... | | Authors: | Rabe, P, Clifton, I, Walla, C, Schofield, C.J. | | Deposit date: | 2021-09-10 | | Release date: | 2022-07-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Spectroscopic studies reveal details of substrate-induced conformational changes distant from the active site in isopenicillin N synthase.
J.Biol.Chem., 298, 2022
|
|
3DTV
 
 | | Crystal structure of arylmalonate decarboxylase | | Descriptor: | Arylmalonate decarboxylase, BETA-MERCAPTOETHANOL, GLYCEROL, ... | | Authors: | Nakasako, M, Obata, R, Miyamaoto, K, Ohta, H. | | Deposit date: | 2008-07-16 | | Release date: | 2009-07-21 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for Inverting the Enantioselectivity of Arylmalonate Decarboxylase Revealed by the Structural Analysis of the Gly74Cys/Cys188Ser Mutant in the Liganded Form
Biochemistry, 49, 2010
|
|
4IMA
 
 | | The structure of C436M-hLPYK in complex with Citrate/Mn/ATP/Fru-1,6-BP | | Descriptor: | 1,2-ETHANEDIOL, 1,6-di-O-phosphono-beta-D-fructofuranose, ADENOSINE, ... | | Authors: | Zhang, B, Holyoak, T, Fenton, A.W, Tang, Q.L, Prasannan, C.B, Deng, J.P. | | Deposit date: | 2013-01-02 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.954 Å) | | Cite: | Energetic Coupling between an Oxidizable Cysteine and the Phosphorylatable N-Terminus of Human Liver Pyruvate Kinase.
Biochemistry, 52, 2013
|
|
6P5Z
 
 | |
7Q5N
 
 | | Crystal structure of Chaetomium thermophilum Ahp1-Urm1 complex | | Descriptor: | Thioredoxin domain-containing protein, Ubiquitin-related modifier 1, ZINC ION | | Authors: | Ravichandran, K.E, Wilk, P, Grudnik, P, Glatt, S. | | Deposit date: | 2021-11-04 | | Release date: | 2022-08-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | E2/E3-independent ubiquitin-like protein conjugation by Urm1 is directly coupled to cysteine persulfidation.
Embo J., 41, 2022
|
|
7Z59
 
 | | SARS-CoV-2 main protease (Mpro) covalently modified with a penicillin derivative | | Descriptor: | (3S)-4-[[2,4-bis(fluoranyl)phenyl]methoxy]-2-methyl-4-oxidanylidene-3-[[(Z)-3-oxidanylidene-2-(2-phenoxyethanoylamino)prop-1-enyl]amino]butane-2-sulfinic acid, 1,2-ETHANEDIOL, 3C-like proteinase nsp5 | | Authors: | Owen, C.D, Malla, T.R, Brewitz, L, Lukacik, P, Strain-Damerell, C, Mikolajek, H, Muntean, D.G, Aslam, H, Salah, E, Tumber, A, Schofield, C.J, Walsh, M.A. | | Deposit date: | 2022-03-08 | | Release date: | 2022-06-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Penicillin Derivatives Inhibit the SARS-CoV-2 Main Protease by Reaction with Its Nucleophilic Cysteine.
J.Med.Chem., 65, 2022
|
|
5FEX
 
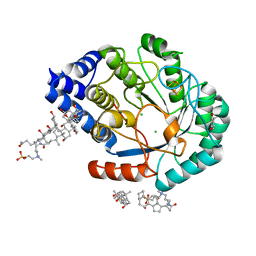 | | HydE from T. maritima in complex with Se-adenosyl-L-selenocysteine (tfinal of the reaction) | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, 5'-DEOXYADENOSINE, CHLORIDE ION, ... | | Authors: | Rohac, R, Amara, P, Benjdia, A, Martin, L, Ruffie, P, Favier, A, Berteau, O, Mouesca, J.M, Fontecilla-Camps, J.C, Nicolet, Y. | | Deposit date: | 2015-12-17 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Carbon-sulfur bond-forming reaction catalysed by the radical SAM enzyme HydE.
Nat.Chem., 8, 2016
|
|
3BB7
 
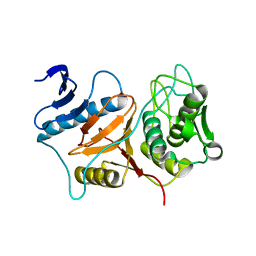 | | Structure of Prevotella intermedia prointerpain A fragment 39-359 (mutant C154A) | | Descriptor: | interpain A | | Authors: | Mallorqui-Fernandez, N, Manandhar, S.P, Mallorqui-Fernandez, G, Uson, I, Wawrzonek, K, Kantyka, T, Sola, M, Thogersen, I.B, Enghild, J.J, Potempa, J, Gomis-Ruth, F.X. | | Deposit date: | 2007-11-09 | | Release date: | 2007-11-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A New Autocatalytic Activation Mechanism for Cysteine Proteases Revealed by Prevotella intermedia Interpain A
J.Biol.Chem., 283, 2008
|
|
4X24
 
 | | Crystal structure of Vibrio cholerae 5'-methylthioadenosine/S-adenosyl homocysteine nucleosidase (MTAN) complexed with methylthio-DADMe-Immucillin-A | | Descriptor: | (3R,4S)-1-[(4-AMINO-5H-PYRROLO[3,2-D]PYRIMIDIN-7-YL)METHYL]-4-[(METHYLSULFANYL)METHYL]PYRROLIDIN-3-OL, 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase, TRIETHYLENE GLYCOL | | Authors: | Cameron, S.A, Thomas, K, Almo, S.C, Schramm, V.L. | | Deposit date: | 2014-11-25 | | Release date: | 2015-08-19 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Active site and remote contributions to catalysis in methylthioadenosine nucleosidases.
Biochemistry, 54, 2015
|
|
6P3O
 
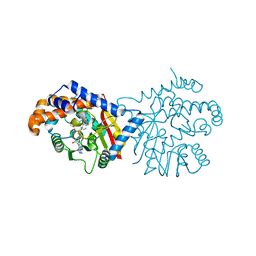 | | Tetrahydroprotoberberine N-methyltransferase in complex with (S)-cis-N-methylstylopine and S-adenosylhomocysteine | | Descriptor: | (5S,12bS)-5-methyl-6,7,12b,13-tetrahydro-2H,4H,10H-[1,3]dioxolo[4,5-g][1,3]dioxolo[7,8]isoquinolino[3,2-a]isoquinolin-5-ium, S-ADENOSYL-L-HOMOCYSTEINE, Tetrahydroprotoberberine N-methyltransferase | | Authors: | Lang, D.E, Morris, J.S, Rowley, M, Torres, M.A, Maksimovich, V.A, Facchini, P.J, Ng, K.K.S. | | Deposit date: | 2019-05-24 | | Release date: | 2019-08-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-function studies of tetrahydroprotoberberineN-methyltransferase reveal the molecular basis of stereoselective substrate recognition.
J.Biol.Chem., 294, 2019
|
|
6H8M
 
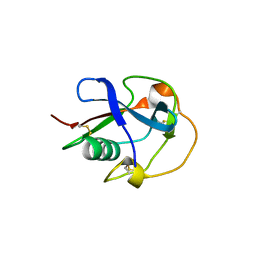 | |
4JYF
 
 | | X-ray snapshots of possible intermediates in the time course of synthesis and degradation of protein-bound Fe4S4 clusters. | | Descriptor: | CARBONATE ION, CHAPSO, CHLORIDE ION, ... | | Authors: | Nicolet, Y, Rohac, R, Martin, L, Fontecilla-Camps, J.C. | | Deposit date: | 2013-03-29 | | Release date: | 2013-05-01 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | X-ray snapshots of possible intermediates in the time course of synthesis and degradation of protein-bound Fe4S4 clusters.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4YML
 
 | | Crystal structure of Escherichia coli 5'-methylthioadenosine/S-adenosyl homocysteine nucleosidase (MTAN) complexed with (3S,4R)-methylthio-DADMe-Immucillin-A | | Descriptor: | (3S,4R)-1-[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]-4-[(methylsulfanyl)methyl]pyrrolidin-3-ol, 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase, PHOSPHATE ION | | Authors: | Cameron, S.A, Thomas, K, Almo, S.C, Schramm, V.L. | | Deposit date: | 2015-03-06 | | Release date: | 2015-08-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Tight binding enantiomers of pre-clinical drug candidates.
Bioorg.Med.Chem., 23, 2015
|
|
