3T3A
 
 | | Crystal structure of H107R mutant of extracellular domain of mouse receptor NKR-P1A | | Descriptor: | Killer cell lectin-like receptor subfamily B member 1A, PHOSPHATE ION | | Authors: | Kolenko, P, Rozbesky, D, Bezouska, K, Hasek, J, Dohnalek, J. | | Deposit date: | 2011-07-25 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the H107R variant of the extracellular domain of mouse NKR-P1A at 2.3 A resolution.
Acta Crystallogr.,Sect.F, 67, 2011
|
|
3QOB
 
 | |
3Q7H
 
 | | Structure of the ClpP subunit of the ATP-dependent Clp Protease from Coxiella burnetii | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, CALCIUM ION, DI(HYDROXYETHYL)ETHER | | Authors: | Anderson, S.M, Wawrzak, Z, Gordon, E, Hasseman, J, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-01-04 | | Release date: | 2011-01-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the ClpP subunit of the ATP-dependent Clp Protease from Coxiella burnetii
To be Published
|
|
3QWC
 
 | | Thrombin Inhibition by Pyridin Derivatives | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, D-phenylalanyl-N-[(4-chloro-1-methylpyridinium-3-yl)methyl]-L-prolinamide, GLYCEROL, ... | | Authors: | Biela, A, Heine, A, Klebe, G. | | Deposit date: | 2011-02-28 | | Release date: | 2012-03-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Impact of ligand and protein desolvation on ligand binding to the S1 pocket of thrombin
J.Mol.Biol., 418, 2012
|
|
3Q5O
 
 | |
3QSE
 
 | | Crystal structure for the complex of substrate-reduced msox with sarcosine | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Monomeric sarcosine oxidase, ... | | Authors: | Kommoju, P, Chen, Z, Bruckner, R.C, Mathews, F.S, Jorns, M.S. | | Deposit date: | 2011-02-21 | | Release date: | 2011-06-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Probing oxygen activation sites in two flavoprotein oxidases using chloride as an oxygen surrogate.
Biochemistry, 50, 2011
|
|
3R2F
 
 | | Crystal structure of beta-site app-cleaving enzyme 1 (BACE-WT) complex with BMS-693391 AKA (2S)-2-((3R)-3-acetamido-3-isobutyl-2-oxo-1-pyrrolidinyl)-N-((1S,2R)-1-(3,5-difluorobenzyl)-2-hydroxy-2-((2R,4R)-4-propoxy-2-pyrrolidinyl)ethyl)-4-phenylbutanamide | | Descriptor: | (2S)-2-[(3R)-3-(acetylamino)-3-(2-methylpropyl)-2-oxopyrrolidin-1-yl]-N-{(1R,2S)-3-(3,5-difluorophenyl)-1-hydroxy-1-[(2R,4R)-4-propoxypyrrolidin-2-yl]propan-2-yl}-4-phenylbutanamide, Beta-secretase 1 | | Authors: | Muckelbauer, J.K. | | Deposit date: | 2011-03-14 | | Release date: | 2011-08-31 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Monosubstituted {gamma}-lactam and conformationally constrained 1,3-diaminopropan-2-ol transition-state isostere inhibitors of {beta}-secretase (BACE).
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3TMZ
 
 | | Crystal Structure of P450 2B4(H226Y) in complex with Amlodipine | | Descriptor: | 5-CYCLOHEXYL-1-PENTYL-BETA-D-MALTOSIDE, Amlodipine, Cytochrome P450 2B4, ... | | Authors: | Shah, M.B, Pascual, J, Stout, C.D, Halpert, J.R. | | Deposit date: | 2011-09-01 | | Release date: | 2012-09-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.248 Å) | | Cite: | Conformational Adaptation of Human Cytochrome P450 2B6 and Rabbit Cytochrome P450 2B4 Revealed upon Binding Multiple Amlodipine Molecules.
Biochemistry, 51, 2012
|
|
3TK3
 
 | | Cytochrome P450 2B4 mutant L437A in complex with 4-(4-chlorophenyl)imidazole | | Descriptor: | 4-(4-CHLOROPHENYL)IMIDAZOLE, Cytochrome P450 2B4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Gay, S.C, Jang, H.H, Wilderman, P.R, Zhang, Q, Stout, C.D, Halpert, J.R. | | Deposit date: | 2011-08-25 | | Release date: | 2011-11-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8001 Å) | | Cite: | Investigation by site-directed mutagenesis of the role of cytochrome P450 2B4 non-active-site residues in protein-ligand interactions based on crystal structures of the ligand-bound enzyme.
Febs J., 279, 2012
|
|
3U4W
 
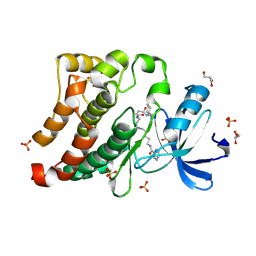 | | Src in complex with DNA-templated macrocyclic inhibitor MC4b | | Descriptor: | GLYCEROL, Proto-oncogene tyrosine-protein kinase Src, SULFATE ION, ... | | Authors: | Seeliger, M.A, Liu, D.R, Georghiou, G, Kleiner, R.E, Pulkoski-Gross, M. | | Deposit date: | 2011-10-10 | | Release date: | 2012-02-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Highly specific, bisubstrate-competitive Src inhibitors from DNA-templated macrocycles.
Nat.Chem.Biol., 8, 2012
|
|
3U8I
 
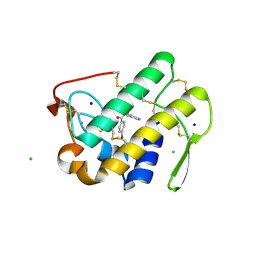 | | Functionally selective inhibition of Group IIA phospholipase A2 reveals a role for vimentin in regulating arachidonic acid metabolism | | Descriptor: | CALCIUM ION, CHLORIDE ION, Phospholipase A2, ... | | Authors: | Lee, L.K, Bryant, K.J, Bouveret, R, Lei, P.-W, Duff, A.P, Harrop, S.J, Huang, E.P, Harvey, R.P, Gelb, M.H, Gray, P.P, Curmi, P.M, Cunningham, A.M, Church, W.B, Scott, K.F. | | Deposit date: | 2011-10-17 | | Release date: | 2012-10-17 | | Last modified: | 2013-06-12 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Selective Inhibition of Human Group IIA-secreted Phospholipase A2 (hGIIA) Signaling Reveals Arachidonic Acid Metabolism Is Associated with Colocalization of hGIIA to Vimentin in Rheumatoid Synoviocytes.
J.Biol.Chem., 288, 2013
|
|
3RE8
 
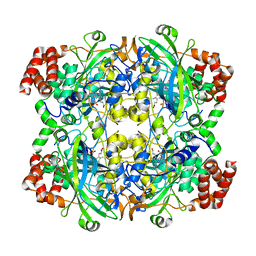 | |
3RJS
 
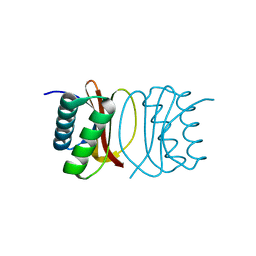 | |
3R1P
 
 | | Odorant Binding Protein 7 from Anopheles gambiae with Four Disulfide Bridges, form P1 | | Descriptor: | Odorant binding protein, antennal, PALMITIC ACID | | Authors: | Lagarde, A, Spinelli, S, Tegoni, M, Field, L, He, X, Zhou, J.J, Cambillau, C. | | Deposit date: | 2011-03-11 | | Release date: | 2011-10-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Crystal Structure of Odorant Binding Protein 7 from Anopheles gambiae Exhibits an Outstanding Adaptability of Its Binding Site.
J.Mol.Biol., 414, 2011
|
|
3SA9
 
 | | Crystal structure of Wild-type HIV-1 protease in complex With AF68 | | Descriptor: | ACETATE ION, N~2~-acetyl-N-[(2S,3R)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}-1-phenylbutan-2-yl]-L-isoleucinamide, PHOSPHATE ION, ... | | Authors: | Schiffer, C.A, Nalam, M.N.L. | | Deposit date: | 2011-06-02 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Protease Inhibitors that protrude out from substrate envelope are more susceptible to developing drug resistance
To be Published
|
|
3SHF
 
 | |
3SEC
 
 | |
3SL8
 
 | | Crystal structure of the catalytic domain of PDE4D2 with compound 10o | | Descriptor: | 1,2-ETHANEDIOL, 3-cyclopentyl 6-ethenyl 2-[(thiophen-2-ylacetyl)amino]-4,7-dihydrothieno[2,3-c]pyridine-3,6(5H)-dicarboxylate, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Feil, S.F. | | Deposit date: | 2011-06-24 | | Release date: | 2011-10-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Thiophene inhibitors of PDE4: Crystal structures show a second binding mode at the catalytic domain of PDE4D2.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3SI0
 
 | | Structure of glycosylated human glutaminyl cyclase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Glutaminyl-peptide cyclotransferase, ... | | Authors: | Parthier, C, Carrillo, D, Stubbs, M.T. | | Deposit date: | 2011-06-17 | | Release date: | 2011-06-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of Glycosylated Mammalian Glutaminyl Cyclases Reveal Conformational Variability near the Active Center.
Biochemistry, 50, 2011
|
|
3S6P
 
 | |
3S9Y
 
 | | Crystal Structure of P. falciparum orotidine 5'-monophosphate decarboxylase complexed with 5-fluoro-6-amino-UMP in space group P21, produced from 5-fluoro-6-azido-UMP | | Descriptor: | 6-amino-5-fluorouridine 5'-(dihydrogen phosphate), DI(HYDROXYETHYL)ETHER, HEXAETHYLENE GLYCOL, ... | | Authors: | Liu, Y, Kotra, L.P, Pai, E.F. | | Deposit date: | 2011-06-02 | | Release date: | 2012-04-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-activity relationships of orotidine-5'-monophosphate decarboxylase inhibitors as anticancer agents.
J.Med.Chem., 52, 2009
|
|
3TS9
 
 | | Crystal Structure of the MDA5 Helicase Insert Domain | | Descriptor: | Interferon-induced helicase C domain-containing protein 1, SULFATE ION | | Authors: | Berke, I.C, Modis, Y. | | Deposit date: | 2011-09-12 | | Release date: | 2012-02-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | MDA5 cooperatively forms dimers and ATP-sensitive filaments upon binding double-stranded RNA.
Embo J., 31, 2012
|
|
3U0Y
 
 | | Crystal structure of the Fucosylgalactoside alpha N-acetylgalactosaminyltransferase (GTA, cisAB mutant L266G, G268A) in complex with compound 382 and UDP | | Descriptor: | 1-(3-phenyl-1,2,4-thiadiazol-5-yl)piperazine, GLYCEROL, Histo-blood group ABO system transferase, ... | | Authors: | Palcic, M.M, Jorgensen, R. | | Deposit date: | 2011-09-29 | | Release date: | 2012-10-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A novel compound from a molecular fragment library screen inhibits glycosyltransferases by displacing the metal ion and interfering with substrate binding
To be Published
|
|
3U8D
 
 | | Functionally selective inhibition of Group IIA phospholipase A2 reveals a role for vimentin in regulating arachidonic acid metabolism | | Descriptor: | (3-{[3-(2-amino-2-oxoethyl)-1-benzyl-2-ethyl-1H-indol-5-yl]oxy}propyl)phosphonic acid, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Lee, L.K, Bryant, K.J, Bouveret, R, Lei, P.-W, Duff, A.P, Harrop, S.J, Huang, E.P, Harvey, R.P, Gelb, M.H, Gray, P.P, Curmi, P.M, Cunningham, A.M, Church, W.B, Scott, K.F. | | Deposit date: | 2011-10-16 | | Release date: | 2012-10-17 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (1.805 Å) | | Cite: | Selective Inhibition of Human Group IIA-secreted Phospholipase A2 (hGIIA) Signaling Reveals Arachidonic Acid Metabolism Is Associated with Colocalization of hGIIA to Vimentin in Rheumatoid Synoviocytes.
J.Biol.Chem., 288, 2013
|
|
3UD2
 
 | | Crystal structure of Selenomethionine ZU5A-ZU5B protein domains of human erythrocyte ankyrin | | Descriptor: | Ankyrin-1, CHLORIDE ION, ETHANOL, ... | | Authors: | Yasunaga, M, Ipsaro, J.J, Mondragon, A. | | Deposit date: | 2011-10-27 | | Release date: | 2012-02-22 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structurally Similar but Functionally Diverse ZU5 Domains in Human Erythrocyte Ankyrin.
J.Mol.Biol., 417, 2012
|
|
