4YTI
 
 | |
5FNU
 
 | | Structure of the Keap1 Kelch domain in complex with a small molecule inhibitor. | | Descriptor: | (3S)-3-(7-methoxy-1-methyl-1H-benzo[d][1,2,3]triazol-5-yl)-3-(4-methyl-3-(((R)-4-methyl-1,1-dioxido-3,4-dihydro-2H-benzo[b][1,4,5]oxathiazepin-2-yl)methyl)phenyl)propanoic acid, CHLORIDE ION, KELCH-LIKE ECH-ASSOCIATED PROTEIN 1 | | Authors: | Davies, T.G, Wixted, W.E, Coyle, J.E, Griffiths-Jones, C, Hearn, K, McMenamin, R, Norton, D, Rich, S.J, Richardson, C, Saxty, G, Willems, H.M.G, Woolford, A.J, Cottom, J.E, Kou, J, Yonchuk, J.G, Feldser, H.G, Sanchez, Y, Foley, J.P, Bolognese, B.J, Logan, G, Podolin, P.L, Yan, H, Callahan, J.F, Heightman, T.D, Kerns, J.K. | | Deposit date: | 2015-11-16 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Mono-Acidic Inhibitors of the Kelch-Like Ech-Associated Protein 1 : Nuclear Factor Erythroid 2-Related Factor 2 (Keap1:Nrf2) Protein-Protein Interaction with High Cell Potency Identified by Fragment-Based Discovery.
J.Med.Chem., 59, 2016
|
|
4DH6
 
 | | Structure of Bace-1 (Beta-Secretase) in Complex with (2R)-N-((2S,3R)-1-(benzo[d][1,3]dioxol-5-yl)-3-hydroxy-4-((S)-6'-neopentyl-3',4'-dihydrospiro[cyclobutane-1,2'-pyrano[2,3-b]pyridine]-4'-ylamino)butan-2-yl)-2-methoxypropanamide | | Descriptor: | (2R)-N-[(2S,3R)-1-(1,3-benzodioxol-5-yl)-4-{[(4'S)-6'-(2,2-dimethylpropyl)-3',4'-dihydrospiro[cyclobutane-1,2'-pyrano[2,3-b]pyridin]-4'-yl]amino}-3-hydroxybutan-2-yl]-2-methoxypropanamide, Beta-secretase 1, GLYCEROL, ... | | Authors: | Sickmier, E.A. | | Deposit date: | 2012-01-27 | | Release date: | 2012-04-18 | | Last modified: | 2013-07-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Design and preparation of a potent series of hydroxyethylamine containing beta-secretase inhibitors that demonstrate robust reduction of central beta-amyloid.
J.Med.Chem., 55, 2012
|
|
5JAX
 
 | | PKG I's Carboyl Terminal Cyclic Nucleotide Binding Domain (CNB-B) in a complex with 8-Br-cGMP | | Descriptor: | 2-amino-8-bromo-9-[(2R,4aR,6R,7R,7aS)-2,7-dihydroxy-2-oxotetrahydro-2H,4H-2lambda~5~-furo[3,2-d][1,3,2]dioxaphosphinin-6-yl]-1,9-dihydro-6H-purin-6-one, CALCIUM ION, SODIUM ION, ... | | Authors: | Campbell, J.C, Sankaran, B, Kim, C.W. | | Deposit date: | 2016-04-12 | | Release date: | 2017-04-19 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.486 Å) | | Cite: | Structural Basis of Analog Specificity in PKG I and II.
ACS Chem. Biol., 12, 2017
|
|
2OOB
 
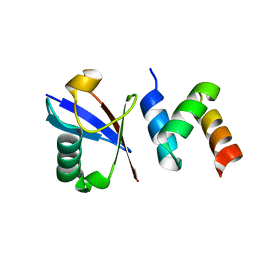 | |
6B8R
 
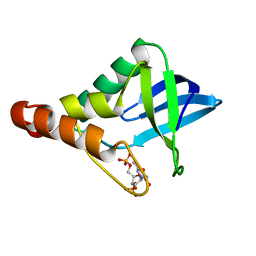 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS V23K/L36Q at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Robinson, A.C, Schlessman, J.L, Garcia-Moreno E, B, Benning, M. | | Deposit date: | 2017-10-09 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Dielectric Properties of a Protein Probed by Reversal of a Buried Ion Pair.
J Phys Chem B, 122, 2018
|
|
3K3A
 
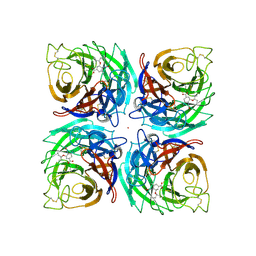 | |
1SGN
 
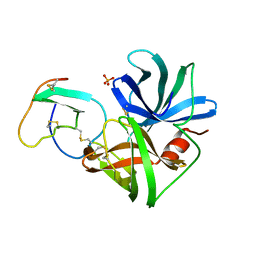 | | ASN 18 VARIANT OF TURKEY OVOMUCOID INHIBITOR THIRD DOMAIN COMPLEXED WITH STREPTOMYCES GRISEUS PROTEINASE B | | Descriptor: | Ovomucoid, PHOSPHATE ION, Streptogrisin B | | Authors: | Huang, K, Lu, W, Anderson, S, Laskowski Jr, M, James, M.N.G. | | Deposit date: | 1999-03-25 | | Release date: | 2003-08-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Recruitment of a Buried K+ Ion to Stabilize the Negative Charge of Ionized P1 in the Hydrophobic Pocket: Crystal Structures of Glu18, Gln18, Asp18 and Asn18 Variants of Turkey Ovomucoid Inhibitor Third Domain Complexed with
Streptomyces griseus Protease B at Various pHs
To be Published
|
|
1SGE
 
 | | GLU 18 VARIANT OF TURKEY OVOMUCOID INHIBITOR THIRD DOMAIN COMPLEXED WITH STREPTOMYCES GRISEUS PROTEINASE B AT PH 6.5 | | Descriptor: | Ovomucoid, PHOSPHATE ION, Streptogrisin B | | Authors: | Huang, K, Lu, W, Anderson, S, Laskowski Jr, M, James, M.N.G. | | Deposit date: | 1999-03-25 | | Release date: | 2003-08-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Recruitment of a Buried K+ Ion to Stabilize the Negative Charge of Ionized P1 in the Hydrophobic Pocket: Crystal Structures of Glu18, Gln18, Asp18 and Asn18 Variants of Turkey Ovomucoid Inhibitor Third Domain Complexed with Streptomyces griseus Protease B at Various pHs
To be Published
|
|
4X8C
 
 | | Crystal structure of human peptidylarginine deiminase type4 (PAD4) in complex with GSK147 | | Descriptor: | CALCIUM ION, Protein-arginine deiminase type-4, [(3S,4R)-3-amino-4-hydroxypiperidin-1-yl]{2-[1-(cyclopropylmethyl)-1H-pyrrolo[2,3-b]pyridin-2-yl]-7-methoxy-1-methyl-1H-benzimidazol-5-yl}methanone | | Authors: | Lewis, H.D, Bax, B.D, Chung, C.-W, Polyakova, O, Thorpe, J. | | Deposit date: | 2014-12-10 | | Release date: | 2015-01-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Inhibition of PAD4 activity is sufficient to disrupt mouse and human NET formation.
Nat.Chem.Biol., 11, 2015
|
|
7WYO
 
 | | Structure of the EV71 3Cpro with 338 inhibitor | | Descriptor: | 3C protein, N-methyl-N-(4,5,6,7-tetrahydro-1,3-benzothiazol-2-ylmethyl)prop-2-enamide | | Authors: | Qin, B, Hou, P, Gao, X, Cui, S. | | Deposit date: | 2022-02-16 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.402 Å) | | Cite: | Acrylamide fragment inhibitors that induce unprecedented conformational distortions in enterovirus 71 3C and SARS-CoV-2 main protease.
Acta Pharm Sin B, 12, 2022
|
|
7WYL
 
 | | Structure of the EV71 3Cpro with 337 inhibitor | | Descriptor: | 3C protein, N-methyl-N-[[4-(trifluoromethyl)-1,3-thiazol-2-yl]methyl]prop-2-enamide | | Authors: | Qin, B, Hou, P, Gao, X, Cui, S. | | Deposit date: | 2022-02-16 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Acrylamide fragment inhibitors that induce unprecedented conformational distortions in enterovirus 71 3C and SARS-CoV-2 main protease.
Acta Pharm Sin B, 12, 2022
|
|
1OJA
 
 | | HUMAN MONOAMINE OXIDASE B IN COMPLEX WITH ISATIN | | Descriptor: | AMINE OXIDASE [FLAVIN-CONTAINING] B, FLAVIN-ADENINE DINUCLEOTIDE, ISATIN | | Authors: | Binda, C, Edmondson, D.E, Mattevi, A. | | Deposit date: | 2003-07-08 | | Release date: | 2003-08-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Insights Into the Mode of Inhibition of Human Mitochondrial Monoamine Oxidase B from High-Resolution Crystal Structures
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
4PCO
 
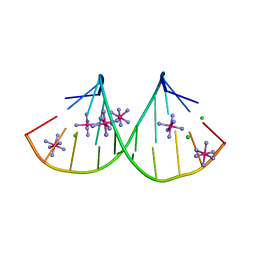 | |
4PO5
 
 | | Crystal structure of allophycocyanin B from Synechocystis PCC 6803 | | Descriptor: | Allophycocyanin beta chain, Allophycocyanin subunit alpha-B, PHYCOCYANOBILIN, ... | | Authors: | Pang, P.P, Dong, L.L, Sun, Y.F, Zeng, X.L, Ding, W.L, Scheer, H, Yang, X, Zhao, K.H. | | Deposit date: | 2014-02-24 | | Release date: | 2014-10-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.751 Å) | | Cite: | The structure of allophycocyanin B from Synechocystis PCC 6803 reveals the structural basis for the extreme redshift of the terminal emitter in phycobilisomes.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5VA4
 
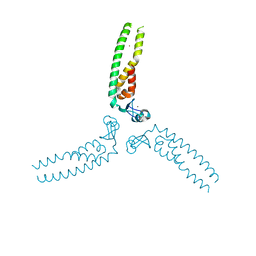 | |
6GG0
 
 | |
1RBB
 
 | |
1I1E
 
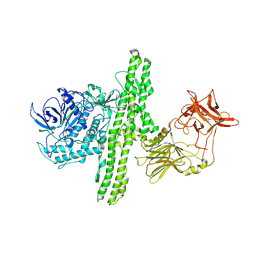 | | CRYSTAL STRUCTURE OF CLOSTRIDIUM BOTULINUM NEUROTOXIN B COMPLEXED WITH DOXORUBICIN | | Descriptor: | BOTULINUM NEUROTOXIN TYPE B, DOXORUBICIN, SULFATE ION, ... | | Authors: | Eswaramoorthy, S, Kumaran, D, Swaminathan, S. | | Deposit date: | 2001-02-01 | | Release date: | 2001-11-21 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystallographic evidence for doxorubicin binding to the receptor-binding site in Clostridium botulinum neurotoxin B.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
4X8G
 
 | | Crystal structure of human peptidylarginine deiminase type4 (PAD4) in complex with GSK199 | | Descriptor: | CALCIUM ION, Protein-arginine deiminase type-4, [(3R)-3-aminopiperidin-1-yl][2-(1-ethyl-1H-pyrrolo[2,3-b]pyridin-2-yl)-7-methoxy-1-methyl-1H-benzimidazol-5-yl]methanone | | Authors: | Lewis, H.D, Bax, B.D, Chung, C.-W, Polyakova, O, Thorpe, J. | | Deposit date: | 2014-12-10 | | Release date: | 2015-01-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Inhibition of PAD4 activity is sufficient to disrupt mouse and human NET formation.
Nat.Chem.Biol., 11, 2015
|
|
5FZ9
 
 | | Crystal structure of the catalytic domain of human JARID1B in complex with Maybridge fragment thieno(3,2-b)thiophene-5-carboxylic acid (N06263b) (ligand modelled based on PANDDA event map, SGC - Diamond I04-1 fragment screening) | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Nowak, R, Krojer, T, Johansson, C, Kupinska, K, Szykowska, A, Pearce, N, Talon, R, Collins, P, Gileadi, C, Strain-Damerell, C, Burgess-Brown, N.A, Arrowsmith, C.H, Bountra, C, Edwards, A.M, von Delft, F, Brennan, P.E, Oppermann, U. | | Deposit date: | 2016-03-12 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal Structure of the Catalytic Domain of Human Jarid1B in Complex with Maybridge Fragment Thieno(3,2-B)Thiophene -5-Carboxylic Acid (N06263B) (Ligand Modelled Based on Pandda Event Map, Sgc - Diamond I04-1 Fragment Screening)
To be Published
|
|
1UYX
 
 | | Carbohydrate binding module (CBM6cm-2) from Cellvibrio mixtus lichenase 5A in complex with cellobiose | | Descriptor: | CALCIUM ION, CELLULASE B, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Czjzek, M, Pires, V.M.R, Henshaw, J, Prates, J.A.M, Bolam, D, Henrissat, B, Gilbert, H.J. | | Deposit date: | 2004-03-03 | | Release date: | 2004-03-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | The Crystal Structure of the Family 6 Carbohydrate Binding Module from Cellvibrio Mixtus Endoglucanase 5A in Complex with Oligosaccharides Reveals Two Distinct Binding Sites with Different Ligand Specificities
J.Biol.Chem., 279, 2004
|
|
1LEC
 
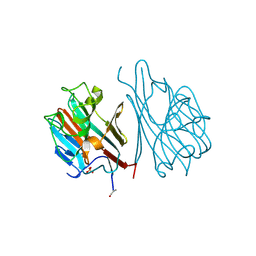 | | STRUCTURES OF THE LECTIN IV OF GRIFFONIA SIMPLICIFOLIA AND ITS COMPLEX WITH THE LEWIS B HUMAN BLOOD GROUP DETERMINANT AT 2.0 ANGSTROMS RESOLUTION | | Descriptor: | CALCIUM ION, MANGANESE (II) ION, SULFATE ION, ... | | Authors: | Delbaere, L, Vandonselaar, M, Quail, J. | | Deposit date: | 1992-12-17 | | Release date: | 1994-01-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of the lectin IV of Griffonia simplicifolia and its complex with the Lewis b human blood group determinant at 2.0 A resolution.
J.Mol.Biol., 230, 1993
|
|
4MJI
 
 | | T cell response to a HIV reverse transcriptase epitope presented by the protective allele HLA-B*51:01 | | Descriptor: | Beta-2-microglobulin, HIV Reverse Transcriptase peptide Marker, HLA class I histocompatibility antigen, ... | | Authors: | Rizkallah, P.J, Cole, D.K, Sewell, A.K, Motozono, C, Takiguchi, M. | | Deposit date: | 2013-09-03 | | Release date: | 2014-05-28 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Molecular basis of a dominant T cell response to an HIV reverse transcriptase 8-mer epitope presented by the protective allele HLA-B*51:01
J.Immunol., 192, 2014
|
|
4N6V
 
 | | Partial rotational order disorder structure of human stefin B | | Descriptor: | Cystatin-B, SULFATE ION | | Authors: | Renko, M, Taler-Vercic, A, Mihelic, M, Zerovnik, E, Turk, D. | | Deposit date: | 2013-10-14 | | Release date: | 2014-04-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Partial rotational lattice order-disorder in stefin B crystals.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
