4LL5
 
 | | Crystal Structure of Pim-1 in complex with the fluorescent compound SKF86002 | | Descriptor: | 6-(4-fluorophenyl)-5-(pyridin-4-yl)-2,3-dihydroimidazo[2,1-b][1,3]thiazole, CALCIUM ION, GLYCEROL, ... | | Authors: | Parker, L.J, Tanaka, A, Handa, N, Honda, K, Tomabechi, Y, Shirouzu, M, Yokoyama, S. | | Deposit date: | 2013-07-09 | | Release date: | 2014-02-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Kinase crystal identification and ATP-competitive inhibitor screening using the fluorescent ligand SKF86002.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4LLI
 
 | |
4LIJ
 
 | |
4LJZ
 
 | | Crystal Structure Analysis of the E.coli holoenzyme | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Bae, B, Darst, S.A. | | Deposit date: | 2013-07-05 | | Release date: | 2013-11-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.5871 Å) | | Cite: | Phage T7 Gp2 inhibition of Escherichia coli RNA polymerase involves misappropriation of sigma 70 domain 1.1.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4LDF
 
 | | Crystal Structure of CpBRD2 from cryptosporidium, cgd3_3190 | | Descriptor: | GCN5 like acetylase + bromodomain, GLYCEROL, UNKNOWN ATOM OR ION | | Authors: | Wernimont, A.K, Loppnau, P, Fonseca, M, Knapp, S, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Hui, R, Mottaghi, K, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-06-24 | | Release date: | 2013-07-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of CpBRD2 from cryptosporidium, cgd3_3190
TO BE PUBLISHED
|
|
4LDZ
 
 | | Crystal structure of the full-length response regulator DesR in the active state | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Trajtenberg, F, Larrieux, N, Buschiazzo, A. | | Deposit date: | 2013-06-25 | | Release date: | 2014-07-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Allosteric activation of bacterial response regulators: the role of the cognate histidine kinase beyond phosphorylation.
MBio, 5, 2014
|
|
4WF7
 
 | | Crystal structures of trehalose synthase from Deinococcus radiodurans reveal that a closed conformation is involved in the intramolecular isomerization catalysis | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Wang, Y.L, Chow, S.Y, Lin, Y.T, Hsieh, Y.C, Lee, G.C, Liaw, S.H. | | Deposit date: | 2014-09-13 | | Release date: | 2014-12-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structures of trehalose synthase from Deinococcus radiodurans reveal that a closed conformation is involved in catalysis of the intramolecular isomerization.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4LEK
 
 | | Structure-Based Design of New Dihydrofolate Reductase Antibacterial Agents: 7-(Benzimidazol-1-yl)-2,4-diaminoquinazolines | | Descriptor: | 7-[5,6-dimethoxy-2-(1,3-thiazol-2-yl)-1H-benzimidazol-1-yl]quinazoline-2,4-diamine, Dihydrofolate reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Hilgers, M.T. | | Deposit date: | 2013-06-25 | | Release date: | 2014-02-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-Based Design of New Dihydrofolate Reductase Antibacterial Agents: 7-(Benzimidazol-1-yl)-2,4-diaminoquinazolines.
J.Med.Chem., 57, 2014
|
|
4WHD
 
 | | Human CEACAM1 N-domain homodimer | | Descriptor: | Carcinoembryonic antigen-related cell adhesion molecule 1, GLYCEROL, octyl beta-D-glucopyranoside | | Authors: | Kirouac, K.N, Prive, G.G. | | Deposit date: | 2014-09-22 | | Release date: | 2015-10-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Human CEACAM1 N-domain homodimer
To Be Published
|
|
4LK9
 
 | | Crystal Structure of MOZ double PHD finger histone H3 tail complex | | Descriptor: | Histone H3.1, Histone acetyltransferase KAT6A, ZINC ION | | Authors: | Dreveny, I, Deeves, S.E, Yue, B, Heery, D.M. | | Deposit date: | 2013-07-07 | | Release date: | 2013-10-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The double PHD finger domain of MOZ/MYST3 induces alpha-helical structure of the histone H3 tail to facilitate acetylation and methylation sampling and modification.
Nucleic Acids Res., 42, 2014
|
|
4LKM
 
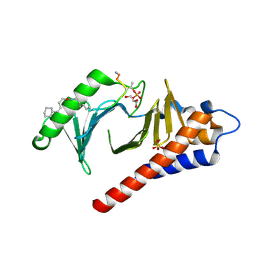 | | Crystal structure of Plk1 Polo-box domain in complex with PL-74 | | Descriptor: | GLYCEROL, PL-74, SULFATE ION, ... | | Authors: | Lee, W.C, Song, J.H, Kim, H.Y. | | Deposit date: | 2013-07-08 | | Release date: | 2013-12-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Exploring the binding nature of pyrrolidine pocket-dependent interactions in the polo-box domain of polo-like kinase 1
Plos One, 8, 2013
|
|
4LKU
 
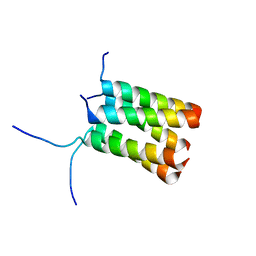 | |
4WPZ
 
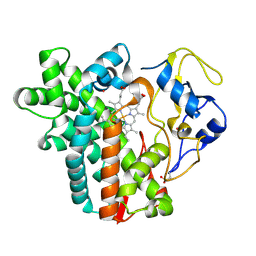 | | Crystal structure of cytochrome P450 CYP107W1 from Streptomyces avermitilis | | Descriptor: | Cytochrome P450, POTASSIUM ION, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Kang, L.W, Kim, D.H, Pham, T.V, Han, S.H. | | Deposit date: | 2014-10-21 | | Release date: | 2015-04-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Functional characterization of CYP107W1 from Streptomyces avermitilis and biosynthesis of macrolide oligomycin A.
Arch.Biochem.Biophys., 575, 2015
|
|
4LH3
 
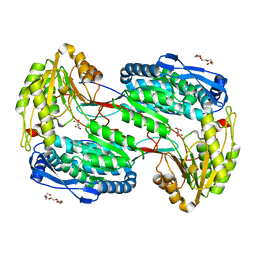 | |
4LKV
 
 | | Determinants of lipid substrate and membrane binding for the tetraacyldisaccharide-1-phosphate 4 -kinase LpxK | | Descriptor: | (R)-((2R,3S,4R,5R,6R)-3-HYDROXY-2-(HYDROXYMETHYL)-5-((R)-3-HYDROXYTETRADECANAMIDO)-6-(PHOSPHONOOXY)TETRAHYDRO-2H-PYRAN-4-YL) 3-HYDROXYTETRADECANOATE, 2-deoxy-3-O-[(3R)-3-hydroxytetradecanoyl]-2-{[(3R)-3-hydroxytetradecanoyl]amino}-4-O-phosphono-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Emptage, R.P, Tonthat, N.K, York, J.D, Schumacher, M.A, Zhou, P. | | Deposit date: | 2013-07-08 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.5109 Å) | | Cite: | Structural Basis of Lipid Binding for the Membrane-embedded Tetraacyldisaccharide-1-phosphate 4'-Kinase LpxK.
J.Biol.Chem., 289, 2014
|
|
4LMI
 
 | |
4LH1
 
 | |
4LJQ
 
 | | Crystal structure of the catalytic core of E3 ligase HOIP | | Descriptor: | E3 ubiquitin-protein ligase RNF31, ZINC ION | | Authors: | Stieglitz, B, Rana, R.R, Koliopoulos, M.G, Morris-Davies, A.C, Christodoulou, E, Howell, S, Brown, N.R, Rittinger, K. | | Deposit date: | 2013-07-05 | | Release date: | 2013-10-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural basis for ligase-specific conjugation of linear ubiquitin chains by HOIP.
Nature, 503, 2013
|
|
4LEH
 
 | |
4LGU
 
 | | Crystal structure of clAP1 BIR3 bound to T3226692 | | Descriptor: | (3S,8aR)-N-((R)-chroman-4-yl)-2-((S)-2-cyclohexyl-2-((S)-2-(methylamino)propanamido)acetyl)octahydropyrrolo[1,2-a]pyrazine-3-carboxamide, Baculoviral IAP repeat-containing protein 2, ZINC ION | | Authors: | Dougan, D.R, Mol, C.D, Snell, G.P. | | Deposit date: | 2013-06-28 | | Release date: | 2013-08-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design, stereoselective synthesis, and biological evaluation of novel tri-cyclic compounds as inhibitor of apoptosis proteins (IAP) antagonists.
Bioorg.Med.Chem., 21, 2013
|
|
4LKL
 
 | |
4LKR
 
 | | Crystal structure of deoD-3 gene product from Shewanella oneidensis MR-1, NYSGRC target 029437 | | Descriptor: | Purine nucleoside phosphorylase DeoD-type | | Authors: | Malashkevich, V.N, Bonanno, J.B, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-07-08 | | Release date: | 2013-07-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of deoD-3 gene product from Shewanella oneidensis MR-1, NYSGRC target 029437
To be Published
|
|
4LK0
 
 | | Crystal Structure Analysis of the E.coli holoenzyme/T7 Gp2 complex | | Descriptor: | Bacterial RNA polymerase inhibitor, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Bae, B, Darst, S.A. | | Deposit date: | 2013-07-05 | | Release date: | 2013-11-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.9103 Å) | | Cite: | Phage T7 Gp2 inhibition of Escherichia coli RNA polymerase involves misappropriation of sigma 70 domain 1.1.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4LOA
 
 | | X-ray structure of the de-novo design amidase at the resolution 1.8A, Northeast Structural Genomics Consortium (NESG) Target OR398 | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, De-novo design amidase | | Authors: | Kuzin, A, Lew, S, Vorobiev, S.M, Seetharaman, J, Sahdev, S, Xiao, R, Maglaqui, M, Kogan, S, Khersonsky, O, Everett, J.K, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-07-12 | | Release date: | 2013-08-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.819 Å) | | Cite: | Northeast Structural Genomics Consortium Target OR398
To be Published
|
|
4WEG
 
 | | influenza virus neuraminidase N9 in complex 2,3-difluorosialic acid | | Descriptor: | (2R,3R,4R,5R,6R)-5-acetamido-2,3-difluoro-4-hydroxy-6-[(1R,2R)-1,2,3-trihydroxypropyl]tetrahydro-2H-pyran-2-carboxylic acid, (3R,4R,5R,6R)-5-(acetylamino)-3-fluoro-4-hydroxy-6-[(1R,2R)-1,2,3-trihydroxypropyl]-3,4,5,6-tetrahydropyranium-2-carboxylate, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Streltsov, V.A, Pilling, P, Barrett, S, McKimm-Breschkin, J. | | Deposit date: | 2014-09-10 | | Release date: | 2015-09-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Catalytic mechanism and novel receptor binding sites of human parainfluenza virus type 3 hemagglutinin-neuraminidase (hPIV3 HN)
Antiviral Res., 123, 2015
|
|
