1II7
 
 | | Crystal structure of P. furiosus Mre11 with manganese and dAMP | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, MANGANESE (II) ION, Mre11 nuclease, ... | | Authors: | Hopfner, K.-P, Karcher, A, Craig, L, Woo, T.T, Carney, J.P, Tainer, J.A. | | Deposit date: | 2001-04-20 | | Release date: | 2001-05-30 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural biochemistry and interaction architecture of the DNA double-strand break repair Mre11 nuclease and Rad50-ATPase.
Cell(Cambridge,Mass.), 105, 2001
|
|
8A8F
 
 | | Crystal structure of Glc7 phosphatase in complex with the regulatory region of Ref2 | | Descriptor: | MANGANESE (II) ION, PHOSPHATE ION, RNA end formation protein 2, ... | | Authors: | Carminati, M, Manav, C.M, Bellini, D, Passmore, L.A. | | Deposit date: | 2022-06-22 | | Release date: | 2023-11-29 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A direct interaction between CPF and RNA Pol II links RNA 3' end processing to transcription.
Mol.Cell, 83, 2023
|
|
6X1Y
 
 | | Mre11 dimer in complex with small molecule modulator PFMI | | Descriptor: | (5Z)-5-[(3-methoxyphenyl)methylidene]-2-sulfanylidene-1,3-thiazolidin-4-one, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Nuclease SbcCD subunit D | | Authors: | Arvai, A.S, Moiani, D, Tainer, J.A. | | Deposit date: | 2020-05-19 | | Release date: | 2020-06-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Fragment- and structure-based drug discovery for developing therapeutic agents targeting the DNA Damage Response.
Prog.Biophys.Mol.Biol., 163, 2021
|
|
7QFB
 
 | | Crystal structure of Protein Phosphatase 1 in complex with PP1-binding peptide from PTG | | Descriptor: | GLYCEROL, MANGANESE (II) ION, Protein phosphatase 1 regulatory subunit 3C, ... | | Authors: | Semrau, M.S, Storici, P, Lolli, G. | | Deposit date: | 2021-12-05 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Molecular architecture of the glycogen- committed PP1/PTG holoenzyme.
Nat Commun, 13, 2022
|
|
7QM2
 
 | |
6XUE
 
 | |
7QWJ
 
 | |
5DNY
 
 | | Structure of the ATPrS-Mre11/Rad50-DNA complex | | Descriptor: | DNA (27-MER), DNA double-strand break repair Rad50 ATPase,DNA double-strand break repair Rad50 ATPase, DNA double-strand break repair protein Mre11, ... | | Authors: | Liu, Y. | | Deposit date: | 2015-09-10 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | ATP-dependent DNA binding, unwinding, and resection by the Mre11/Rad50 complex.
Embo J., 35, 2016
|
|
6XUG
 
 | |
6XUQ
 
 | |
5EBB
 
 | | Structure of human sphingomyelinase phosphodiesterase like 3A (SMPDL3A) with Zn2+ | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Acid sphingomyelinase-like phosphodiesterase 3a, GLYCEROL, ... | | Authors: | Lim, S.M, Yeung, K, Tresaugues, L, Teo, H.L, Nordlund, P. | | Deposit date: | 2015-10-19 | | Release date: | 2016-01-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The structure and catalytic mechanism of human sphingomyelin phosphodiesterase like 3a - an acid sphingomyelinase homologue with a novel nucleotide hydrolase activity.
Febs J., 283, 2016
|
|
8BAH
 
 | | Human Mre11-Nbs1 complex | | Descriptor: | Double-strand break repair protein MRE11, MANGANESE (II) ION, Nibrin | | Authors: | Bartho, J.D, Rotheneder, M, Stakyte, K, Lammens, K, Hopfner, K.P. | | Deposit date: | 2022-10-11 | | Release date: | 2023-01-11 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (4.13 Å) | | Cite: | Cryo-EM structure of the Mre11-Rad50-Nbs1 complex reveals the molecular mechanism of scaffolding functions.
Mol.Cell, 83, 2023
|
|
2BDX
 
 | | X-ray Crystal Structure of dihydromicrocystin-LA bound to Protein Phosphatase-1 | | Descriptor: | DIHYDROMICROCYSTIN-LA, MANGANESE (II) ION, Serine/threonine protein phosphatase PP1-gamma catalytic subunit | | Authors: | Maynes, J.T, Luu, H.A, Cherney, M.M, Andersen, R.J, Williams, D, Holmes, C.F, James, M.N. | | Deposit date: | 2005-10-21 | | Release date: | 2006-01-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of Protein Phosphatase-1 Bound to Motuporin and Dihydromicrocystin-LA: Elucidation of the Mechanism of Enzyme Inhibition by Cyanobacterial Toxins.
J.Mol.Biol., 356, 2006
|
|
8BRN
 
 | | Crystal structure of red kidney bean purple acid phosphatase in complex with an alpha-aminonaphthylmethylphosphonic acid inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Feder, D, Hussein, W.M, Guddat, L.W, Schenk, G. | | Deposit date: | 2022-11-23 | | Release date: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Optimization of an alpha-aminonaphthylmethylphosphonic acid inhibitor of purple acid phosphatase using rational structure-based design approaches.
Eur.J.Med.Chem., 254, 2023
|
|
2IE3
 
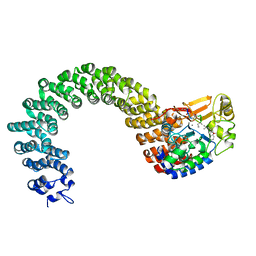 | | Structure of the Protein Phosphatase 2A Core Enzyme Bound to Tumor-inducing Toxins | | Descriptor: | MANGANESE (II) ION, Protein Phosphatase 2, regulatory subunit A (PR 65), ... | | Authors: | Xing, Y, Xu, Y, Chen, Y, Jeffrey, P.D, Chao, Y, Shi, Y. | | Deposit date: | 2006-09-17 | | Release date: | 2006-11-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of Protein Phosphatase 2A Core Enzyme Bound to Tumor-Inducing Toxins
Cell(Cambridge,Mass.), 127, 2006
|
|
1JK7
 
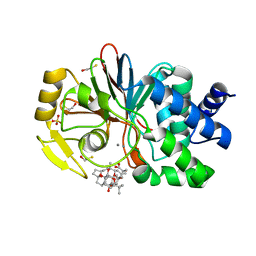 | | CRYSTAL STRUCTURE OF THE TUMOR-PROMOTER OKADAIC ACID BOUND TO PROTEIN PHOSPHATASE-1 | | Descriptor: | BETA-MERCAPTOETHANOL, MANGANESE (II) ION, OKADAIC ACID, ... | | Authors: | Maynes, J.T, Bateman, K.S, Cherney, M.M, Das, A.K, James, M.N. | | Deposit date: | 2001-07-11 | | Release date: | 2001-08-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the tumor-promoter okadaic acid bound to protein phosphatase-1.
J.Biol.Chem., 276, 2001
|
|
2IAE
 
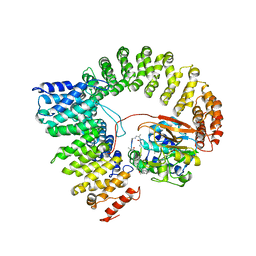 | | Crystal structure of a protein phosphatase 2A (PP2A) holoenzyme. | | Descriptor: | MANGANESE (II) ION, Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit gamma isoform, Serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A alpha isoform, ... | | Authors: | Cho, U.S, Xu, W. | | Deposit date: | 2006-09-07 | | Release date: | 2006-12-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of a protein phosphatase 2A heterotrimeric holoenzyme.
Nature, 445, 2007
|
|
1HP1
 
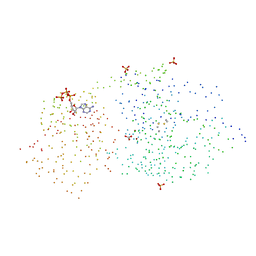 | | 5'-NUCLEOTIDASE (OPEN FORM) COMPLEX WITH ATP | | Descriptor: | 5'-NUCLEOTIDASE, ADENOSINE-5'-TRIPHOSPHATE, CARBONATE ION, ... | | Authors: | Knoefel, T, Straeter, N. | | Deposit date: | 2000-12-12 | | Release date: | 2002-03-20 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism of hydrolysis of phosphate esters by the dimetal center of 5'-nucleotidase based on crystal structures.
J.Mol.Biol., 309, 2001
|
|
5EBE
 
 | | Structure of human sphingomyelinase phosphodiesterase like 3A (SMPDL3A) with 5' CMP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5-O-phosphono-beta-D-ribofuranose, ... | | Authors: | Lim, S.M, Yeung, K, Tresaugues, L, Teo, H.L, Nordlund, P. | | Deposit date: | 2015-10-19 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structure and catalytic mechanism of human sphingomyelin phosphodiesterase like 3a - an acid sphingomyelinase homologue with a novel nucleotide hydrolase activity.
Febs J., 283, 2016
|
|
6S6V
 
 | | Resting state of the E. coli Mre11-Rad50 (SbcCD) head complex bound to ATPgS | | Descriptor: | MAGNESIUM ION, MANGANESE (II) ION, Nuclease SbcCD subunit C, ... | | Authors: | Kaeshammer, L, Saathoff, J.H, Gut, F, Bartho, J, Alt, A, Kessler, B, Lammens, K, Hopfner, K.P. | | Deposit date: | 2019-07-03 | | Release date: | 2019-09-04 | | Last modified: | 2019-11-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Mechanism of DNA End Sensing and Processing by the Mre11-Rad50 Complex.
Mol.Cell, 76, 2019
|
|
6YE1
 
 | | Human Ecto-5'-nucleotidase (CD73) in complex with the AMPCP derivative A894 (compound 2n in publication) in the closed form (crystal form IV) | | Descriptor: | 5'-nucleotidase, ZINC ION, [(2~{R},3~{S},4~{R},5~{R})-5-[2-chloranyl-6-(cyclopentylamino)purin-9-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methoxymethylphosphonic acid | | Authors: | Scaletti, E, Strater, N. | | Deposit date: | 2020-03-23 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Discovery of Potent and Selective Methylenephosphonic Acid CD73 Inhibitors.
J.Med.Chem., 64, 2021
|
|
1KBP
 
 | | KIDNEY BEAN PURPLE ACID PHOSPHATASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FE (III) ION, PURPLE ACID PHOSPHATASE, ... | | Authors: | Klabunde, T, Strater, N, Krebs, B. | | Deposit date: | 1995-02-20 | | Release date: | 1996-10-14 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Mechanism of Fe(III)-Zn(II) purple acid phosphatase based on crystal structures.
J.Mol.Biol., 259, 1996
|
|
6S85
 
 | | Cutting state of the E. coli Mre11-Rad50 (SbcCD) head complex bound to ADP and dsDNA. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (31-MER), DNA (32-MER), ... | | Authors: | Kaeshammer, L, Saathoff, J.H, Gut, F, Bartho, J, Alt, A, Kessler, B, Lammens, K, Hopfner, K.P. | | Deposit date: | 2019-07-08 | | Release date: | 2019-09-04 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Mechanism of DNA End Sensing and Processing by the Mre11-Rad50 Complex.
Mol.Cell, 76, 2019
|
|
6S7H
 
 | | Human CD73 (5'-nucleotidase) in complex with PSB12489 (an AOPCP derivative) in the closed state | | Descriptor: | (N6,N6)-methyl,benzyl-C2-chloro-(alpha,beta)-methylene-ADP, 5'-nucleotidase, CALCIUM ION, ... | | Authors: | Pippel, J, Strater, N. | | Deposit date: | 2019-07-04 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | X-Ray Co-Crystal Structure Guides the Way to Subnanomolar Competitive Ecto-5'-Nucleotidase (CD73) Inhibitors for Cancer Immunotherapy
Adv Ther, 2019
|
|
2IE4
 
 | | Structure of the Protein Phosphatase 2A Core Enzyme Bound to okadaic acid | | Descriptor: | MANGANESE (II) ION, OKADAIC ACID, Protein Phosphatase 2, ... | | Authors: | Xing, Y, Xu, Y, Chen, Y, Jeffrey, P.D, Chao, Y, Shi, Y. | | Deposit date: | 2006-09-17 | | Release date: | 2006-11-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of Protein Phosphatase 2A Core Enzyme Bound to Tumor-Inducing Toxins
Cell(Cambridge,Mass.), 127, 2006
|
|
