6NIP
 
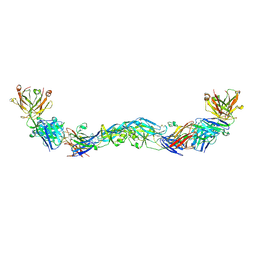 | | Crystal structure of a human anti-ZIKV-DENV neutralizing antibody MZ1 in complex with ZIKV E glycoprotein | | Descriptor: | Envelope protein E, MZ1 Heavy chain, MZ1 Light Chain | | Authors: | Sankhala, R.S, Dussupt, V, Donofrio, G, Choe, M, Modjarrad, K, Michael, N.L, Krebs, S.J, Joyce, M.G. | | Deposit date: | 2018-12-31 | | Release date: | 2019-12-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (4.16 Å) | | Cite: | Potent Zika and dengue cross-neutralizing antibodies induced by Zika vaccination in a dengue-experienced donor.
Nat Med, 26, 2020
|
|
7TY3
 
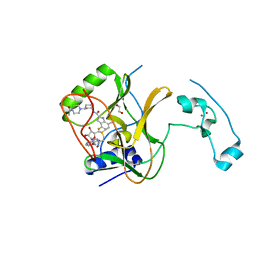 | | Crystal Structure of SETD2 Bound to an Indole-based Inhibitor | | Descriptor: | 1,2-ETHANEDIOL, Histone-lysine N-methyltransferase SETD2, N-[(1R,3R)-3-(4-acetylpiperazin-1-yl)cyclohexyl]-4-fluoro-7-methyl-1H-indole-2-carboxamide, ... | | Authors: | Farrow, N.A. | | Deposit date: | 2022-02-11 | | Release date: | 2022-08-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Conformational-Design-Driven Discovery of EZM0414: A Selective, Potent SETD2 Inhibitor for Clinical Studies.
Acs Med.Chem.Lett., 13, 2022
|
|
7TY2
 
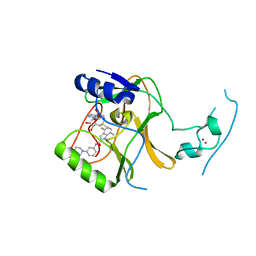 | | Crystal Structure of SETD2 Bound to an Indole-based Inhibitor | | Descriptor: | Histone-lysine N-methyltransferase SETD2, N-[(1R,3S)-3-(4-acetylpiperazin-1-yl)cyclohexyl]-4-fluoro-7-methyl-1H-indole-2-carboxamide, S-ADENOSYLMETHIONINE, ... | | Authors: | Farrow, N.A. | | Deposit date: | 2022-02-11 | | Release date: | 2022-08-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.438 Å) | | Cite: | Conformational-Design-Driven Discovery of EZM0414: A Selective, Potent SETD2 Inhibitor for Clinical Studies.
Acs Med.Chem.Lett., 13, 2022
|
|
5TKJ
 
 | | Structure of vaccine-elicited diverse HIV-1 neutralizing antibody vFP1.01 in complex with HIV-1 fusion peptide residue 512-519 | | Descriptor: | HIV-1 fusion peptide residue 512-519, SULFATE ION, vFP1.01 chimeric mouse antibody heavy chain, ... | | Authors: | Xu, K, Liu, K, Kwong, P.D. | | Deposit date: | 2016-10-06 | | Release date: | 2018-04-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.118 Å) | | Cite: | Epitope-based vaccine design yields fusion peptide-directed antibodies that neutralize diverse strains of HIV-1.
Nat. Med., 24, 2018
|
|
7JIS
 
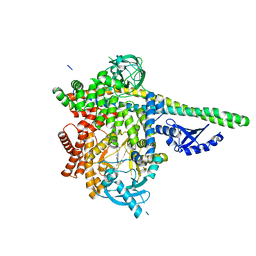 | | HUMAN PI3KDELTA IN COMPLEX WITH COMPOUND 2F | | Descriptor: | (3S)-3-benzyl-5-[9-ethyl-8-(2-methylpyrimidin-5-yl)-9H-purin-6-yl]-3-methyl-1,3-dihydro-2H-indol-2-one, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Lesburg, C.A, Augustin, M. | | Deposit date: | 2020-07-23 | | Release date: | 2020-12-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Optimization of Versatile Oxindoles as Selective PI3K delta Inhibitors.
Acs Med.Chem.Lett., 11, 2020
|
|
1BQ1
 
 | | E. COLI THYMIDYLATE SYNTHASE MUTANT N177A IN COMPLEX WITH CB3717 AND 2'-DEOXYURIDINE 5'-MONOPHOSPHATE (DUMP) | | Descriptor: | 10-PROPARGYL-5,8-DIDEAZAFOLIC ACID, 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, THYMIDYLATE SYNTHASE | | Authors: | Reyes, C.L, Sage, C.R, Rutenber, E.E, Finer-Moore, J.S, Stroud, R.M. | | Deposit date: | 1998-08-20 | | Release date: | 1999-05-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Inactivity of N229A thymidylate synthase due to water-mediated effects: isolating a late stage in methyl transfer.
J.Mol.Biol., 284, 1998
|
|
7W3L
 
 | | Crystal structure of LSD1 in complex with cis-4-Br-2,5-F2-PCPA (S1024) | | Descriptor: | 3-[4-bromanyl-2,5-bis(fluoranyl)phenyl]propanal, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Niwa, H, Sato, S, Umehara, T. | | Deposit date: | 2021-11-25 | | Release date: | 2022-09-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structure-Activity Relationship and In Silico Evaluation of cis- and trans-PCPA-Derived Inhibitors of LSD1 and LSD2
Acs Med.Chem.Lett., 13, 2022
|
|
8SVY
 
 | | MBP-Mcl1 in complex with ligand 10 | | Descriptor: | (15P)-17-chloro-33-fluoro-12-[(2-methoxyethoxy)methyl]-5,14,22-trimethyl-28-oxa-9-thia-5,6,13,14,22-pentaazaheptacyclo[27.7.1.1~4,7~.0~11,15~.0~16,21~.0~20,24~.0~30,35~]octatriaconta-1(36),4(38),6,11(15),12,16,18,20,23,29(37),30,32,34-tridecaene-23-carboxylic acid, 1,2-ETHANEDIOL, GLYCEROL, ... | | Authors: | Miller, B.R, Shaffer, P. | | Deposit date: | 2023-05-17 | | Release date: | 2023-08-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Macrocyclic Carbon-Linked Pyrazoles As Novel Inhibitors of MCL-1.
Acs Med.Chem.Lett., 14, 2023
|
|
5UOP
 
 | | CRYSTAL STRUCTURE OF THE PROTOTYPE FOAMY VIRUS INTASOME WITH A 2- PYRIDINONE AMINAL INHIBITOR (COMPOUND 18) | | Descriptor: | (1S,2S,5R)-8'-[(3-chloro-4-fluorophenyl)methyl]-2'-[2-(2,5-dioxo-2,5-dihydro-1H-pyrrol-1-yl)ethyl]-6'-hydroxy-9',10'-dihydro-2'H-spiro[bicyclo[3.1.0]hexane-2,3'-imidazo[5,1-a][2,6]naphthyridine]-1',5',7'(8'H)-trione, GLYCEROL, INTEGRASE, ... | | Authors: | Klein, D.J. | | Deposit date: | 2017-02-01 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Discovery and optimization of 2-pyridinone aminal integrase strand transfer inhibitors for the treatment of HIV.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
6OUV
 
 | |
7F5S
 
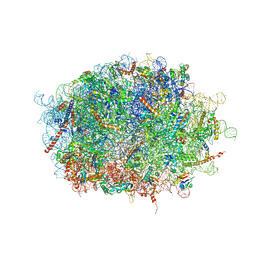 | | human delta-METTL18 60S ribosome | | Descriptor: | 28S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Takahashi, M, Kashiwagi, K, Ito, T. | | Deposit date: | 2021-06-22 | | Release date: | 2022-06-22 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | METTL18-mediated histidine methylation of RPL3 modulates translation elongation for proteostasis maintenance.
Elife, 11, 2022
|
|
7UNF
 
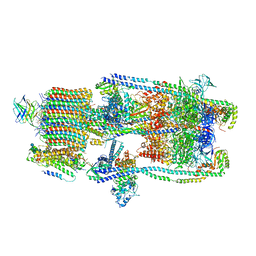 | | CryoEM structure of a mEAK7 bound human V-ATPase complex | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Wang, R, Li, X. | | Deposit date: | 2022-04-10 | | Release date: | 2022-06-15 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.08 Å) | | Cite: | Molecular basis of mEAK7-mediated human V-ATPase regulation.
Nat Commun, 13, 2022
|
|
8TTY
 
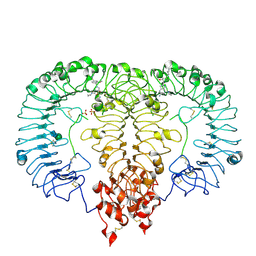 | | Crystal structure of monkey TLR7 ectodomain with compound 5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, N~7~-butyl-2-({4-[(cyclobutylamino)methyl]-2-methoxyphenyl}methyl)-2H-pyrazolo[4,3-d]pyrimidine-5,7-diamine, ... | | Authors: | Critton, D.A. | | Deposit date: | 2023-08-15 | | Release date: | 2024-02-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Discovery of Novel TLR7 Agonists as Systemic Agent for Combination With aPD1 for Use in Immuno-oncology.
Acs Med.Chem.Lett., 15, 2024
|
|
8TTZ
 
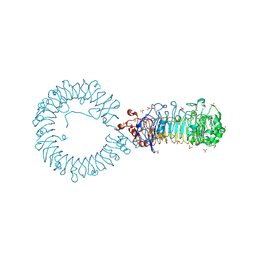 | | Crystal structure of monkey TLR7 ectodomain with compound 20 | | Descriptor: | (3S)-3-({5-amino-1-[(2-methoxy-4-{[(oxan-4-yl)amino]methyl}phenyl)methyl]-1H-pyrazolo[4,3-d]pyrimidin-7-yl}amino)hexan-1-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Critton, D.A. | | Deposit date: | 2023-08-15 | | Release date: | 2024-02-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.14 Å) | | Cite: | Discovery of Novel TLR7 Agonists as Systemic Agent for Combination With aPD1 for Use in Immuno-oncology.
Acs Med.Chem.Lett., 15, 2024
|
|
5UOQ
 
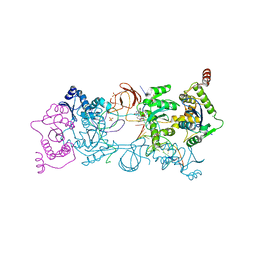 | | CRYSTAL STRUCTURE OF THE PROTOTYPE FOAMY VIRUS INTASOME WITH A 2- PYRIDINONE AMINAL INHIBITOR (COMPOUND 31) | | Descriptor: | (3R)-8-[(3-chloro-4-fluorophenyl)methyl]-6-hydroxy-1,5,7-trioxo-1,2',3',5,7,8,9,10-octahydro-2H-spiro[imidazo[5,1-a][2,6]naphthyridine-3,1'-indene]-7'-carbonitrile, GLYCEROL, INTEGRASE, ... | | Authors: | Klein, D.J. | | Deposit date: | 2017-02-01 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Discovery and optimization of 2-pyridinone aminal integrase strand transfer inhibitors for the treatment of HIV.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
7LLB
 
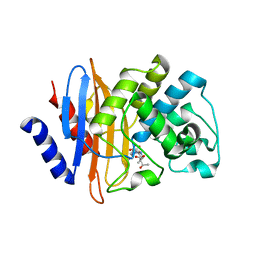 | | Crystal structure of KPC-2 S70G/T215P mutant with hydrolyzed meropenem | | Descriptor: | (2S,3R)-2-[(2S,3R)-1,3-bis(oxidanyl)-1-oxidanylidene-butan-2-yl]-4-[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfan yl-3-methyl-2,3-dihydro-1H-pyrrole-5-carboxylic acid, Carbapenem-hydrolyzing beta-lactamase KPC | | Authors: | Furey, I, Palzkill, T, Sankaran, B, Hu, L, Prasad, B.V.V. | | Deposit date: | 2021-02-03 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Local interactions with the Glu166 base and the conformation of an active site loop play key roles in carbapenem hydrolysis by the KPC-2 beta-lactamase.
J.Biol.Chem., 296, 2021
|
|
7LJK
 
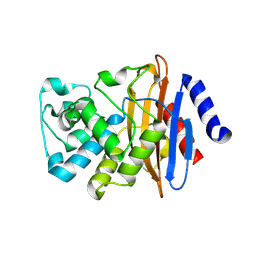 | | Crystal structure of the deacylation deficient KPC-2 F72Y mutant | | Descriptor: | Beta-lactamase | | Authors: | Furey, I, Palzkill, T, Sankaran, B, Hu, L, Prasad, B.V.V. | | Deposit date: | 2021-01-29 | | Release date: | 2021-05-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Local interactions with the Glu166 base and the conformation of an active site loop play key roles in carbapenem hydrolysis by the KPC-2 beta-lactamase.
J.Biol.Chem., 296, 2021
|
|
7LLH
 
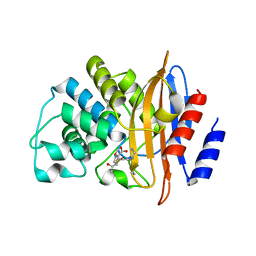 | | KPC-2 F72Y mutant with acylated imipenem | | Descriptor: | (5R)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-3-[(2-{[(E)-iminomethyl]amino}ethyl)sulfanyl]-4,5-dihydro-1H-pyrrole-2-carbox ylic acid, Carbapenem-hydrolyzing beta-lactamase KPC | | Authors: | Furey, I, Palzkill, T, Sankaran, B, Hu, L, Prasad, B.V.V. | | Deposit date: | 2021-02-03 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Local interactions with the Glu166 base and the conformation of an active site loop play key roles in carbapenem hydrolysis by the KPC-2 beta-lactamase.
J.Biol.Chem., 296, 2021
|
|
8DJV
 
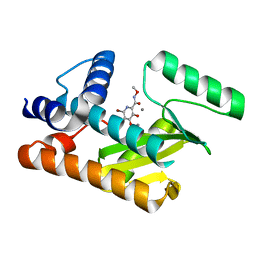 | | The N-terminal domain of PA endonuclease from the influenza H1N1 viral polymerase in complex with 6-Bromo-3-hydroxy-N-methoxy-4-oxo-1,4-dihydropyridine-2-carboxamide | | Descriptor: | 6-bromo-3-hydroxy-N-methoxy-4-oxo-1,4-dihydropyridine-2-carboxamide, MANGANESE (II) ION, Polymerase acidic protein | | Authors: | Kohlbrand, A.J, Stokes, R.W, Karges, J, Seo, H, Sankaran, B, Cohen, S.M. | | Deposit date: | 2022-07-01 | | Release date: | 2022-12-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Carboxylic Acid Isostere Derivatives of Hydroxypyridinones as Core Scaffolds for Influenza Endonuclease Inhibitors.
Acs Med.Chem.Lett., 14, 2023
|
|
4YI5
 
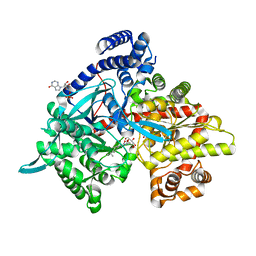 | | Crystal structure of Gpb in complex with 4b | | Descriptor: | Glycogen phosphorylase, muscle form, INOSINIC ACID, ... | | Authors: | Kantsadi, A.L, Chatzileontiadou, D.S.M, Stravodimos, G.A, Leonidas, D.D. | | Deposit date: | 2015-02-27 | | Release date: | 2015-11-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Glycogen phosphorylase as a target for type 2 diabetes: synthetic, biochemical, structural and computational evaluation of novel N-acyl-N -( beta-D-glucopyranosyl) urea inhibitors.
Curr Top Med Chem, 15, 2015
|
|
4TMP
 
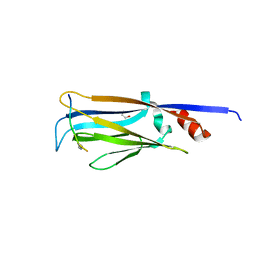 | | Crystal structure of AF9 YEATS bound to H3K9ac peptide | | Descriptor: | 1,2-ETHANEDIOL, ALA-ARG-THR-LYS-GLN-THR-ALA-ARG-ALY-SER-THR, Protein AF-9 | | Authors: | Li, H, Li, Y, Wang, H, Ren, Y. | | Deposit date: | 2014-06-02 | | Release date: | 2014-11-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | AF9 YEATS Domain Links Histone Acetylation to DOT1L-Mediated H3K79 Methylation.
Cell, 159, 2014
|
|
7SJS
 
 | |
1DAE
 
 | | DETHIOBIOTIN SYNTHETASE COMPLEXED WITH 3-(1-AMINOETHYL) NONANEDIOIC ACID | | Descriptor: | 3-(1-AMINOETHYL)NONANEDIOIC ACID, DETHIOBIOTIN SYNTHETASE | | Authors: | Huang, W, Jia, J, Schneider, G, Lindqvist, Y. | | Deposit date: | 1995-05-08 | | Release date: | 1996-06-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism of an ATP-dependent carboxylase, dethiobiotin synthetase, based on crystallographic studies of complexes with substrates and a reaction intermediate.
Biochemistry, 34, 1995
|
|
7SF1
 
 | | SARS-CoV-2 Main Protease (Mpro) in Complex with ML1001 | | Descriptor: | (1R,2S,5S)-N-{(2S,3R)-4-amino-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-3-[N-(3,3-dimethylbutanoyl)-3-methyl-L-valyl]-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Westberg, M, Fernandez, D, Lin, M.Z. | | Deposit date: | 2021-10-02 | | Release date: | 2022-10-05 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | An orally bioavailable SARS-CoV-2 main protease inhibitor exhibits improved affinity and reduced sensitivity to mutations.
Sci Transl Med, 16, 2024
|
|
8PD6
 
 | | Crystal structure of the TRIM58 PRY-SPRY domain in complex with TRIM-473 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, E3 ubiquitin-protein ligase TRIM58, ... | | Authors: | Renatus, M, Hoegenauer, K, Schroeder, M. | | Deposit date: | 2023-06-11 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Discovery of Ligands for TRIM58, a Novel Tissue-Selective E3 Ligase.
Acs Med.Chem.Lett., 14, 2023
|
|
