3LZU
 
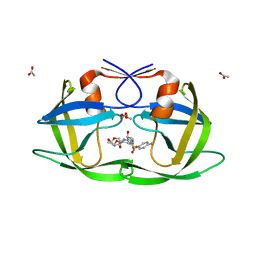 | | Crystal Structure of a Nelfinavir Resistant HIV-1 CRF01_AE Protease variant (N88S) in Complex with the Protease Inhibitor Darunavir. | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, ACETATE ION, HIV-1 protease | | Authors: | Schiffer, C.A, Bandaranayake, R.M. | | Deposit date: | 2010-03-01 | | Release date: | 2010-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | The Effect of Clade-Specific Sequence Polymorphisms on HIV-1 Protease Activity and Inhibitor Resistance Pathways.
J.Virol., 84, 2010
|
|
1AJK
 
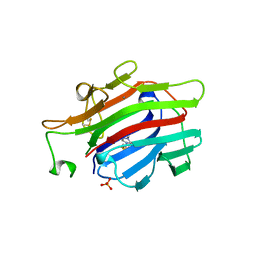 | | CIRCULARLY PERMUTED (1-3,1-4)-BETA-D-GLUCAN 4-GLUCANOHYDROLASE CPA16M-84 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, CIRCULARLY PERMUTED (1-3,1-4)-BETA-D-GLUCAN 4-GLUCANOHYDROLASE, ... | | Authors: | Ay, J, Heinemann, U. | | Deposit date: | 1997-05-06 | | Release date: | 1998-05-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures and properties of de novo circularly permuted 1,3-1,4-beta-glucanases.
Proteins, 30, 1998
|
|
5IRW
 
 | |
3MMF
 
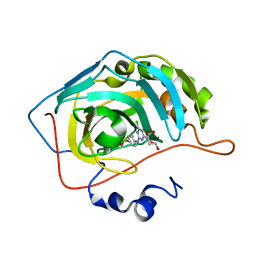 | | Crystal structure of human carbonic anhydrase II in complex with a 1,3,5-triazine-substituted benzenesulfonamide inhibitor | | Descriptor: | 4-({4-chloro-6-[(2-hydroxyethyl)amino]-1,3,5-triazin-2-yl}amino)benzenesulfonamide, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Avvaru, B.S, Wagner, J, Robbins, A.H, Mckenna, R. | | Deposit date: | 2010-04-19 | | Release date: | 2011-04-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Sulfonamides incorporating 1,3,5-triazine moieties selectively and potently inhibit carbonic anhydrase transmembrane isoforms IX, XII and XIV over cytosolic isoforms I and II: Solution and X-ray crystallographic studies.
Bioorg.Med.Chem., 19, 2011
|
|
3MMT
 
 | |
8RBD
 
 | | Crystal structure of Mycobacterium tuberculosis MmaA1 with Sinefungin (SFG) | | Descriptor: | 1,2-ETHANEDIOL, Mycolic acid methyltransferase MmaA1, SINEFUNGIN, ... | | Authors: | Kobakhidze, G, Wachelder, L, Chaudhary, B, Mazumdar, P.A, Madhurantakam, C, Dong, G. | | Deposit date: | 2023-12-04 | | Release date: | 2024-12-11 | | Last modified: | 2025-06-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structures of the mycolic acid methyl transferase 1 (MmaA1) from Mycobacterium tuberculosis in the apo-form and in complex with different cofactors reveal unique features for substrate binding.
J.Biomol.Struct.Dyn., 2025
|
|
3LZV
 
 | | Structure of Nelfinavir-resistant HIV-1 protease (D30N/N88D) in complex with Darunavir. | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, ACETATE ION, HIV-1 Protease, ... | | Authors: | Schiffer, C.A, Kolli, M. | | Deposit date: | 2010-03-01 | | Release date: | 2010-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The Effect of Clade-Specific Sequence Polymorphisms on HIV-1 Protease Activity and Inhibitor Resistance Pathways.
J.Virol., 84, 2010
|
|
7TY7
 
 | | Cryo-EM structure of human Anion Exchanger 1 bound to Bicarbonate | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BICARBONATE ION, ... | | Authors: | Capper, M.J, Mathiharan, Y.K, Yang, S, Stone, A.C, Wacker, D. | | Deposit date: | 2022-02-11 | | Release date: | 2023-08-16 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Substrate binding and inhibition of the anion exchanger 1 transporter.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7TY6
 
 | | Cryo-EM structure of human Anion Exchanger 1 bound to 4,4'-Diisothiocyanatodihydrostilbene-2,2'-Disulfonic Acid (H2DIDS) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4,4'-Diisothiocyano-2,2'-stilbenedisulfonic acid, ... | | Authors: | Capper, M.J, Mathiharan, Y.K, Yang, S, Stone, A.C, Wacker, D. | | Deposit date: | 2022-02-11 | | Release date: | 2023-08-16 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Substrate binding and inhibition of the anion exchanger 1 transporter.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7TY4
 
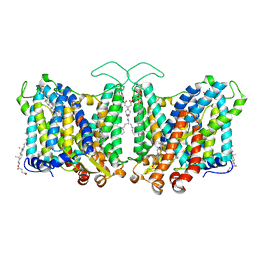 | | Cryo-EM structure of human Anion Exchanger 1 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Band 3 anion transport protein, ... | | Authors: | Capper, M.J, Mathiharan, Y.K, Yang, S, Stone, A.C, Wacker, D. | | Deposit date: | 2022-02-11 | | Release date: | 2023-08-16 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Substrate binding and inhibition of the anion exchanger 1 transporter.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7TYA
 
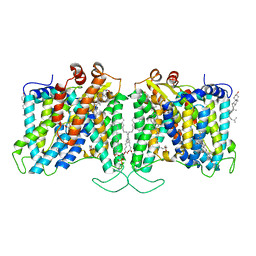 | | Cryo-EM structure of human Anion Exchanger 1 modified with Diethyl Pyrocarbonate (DEPC) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Band 3 anion transport protein, ... | | Authors: | Capper, M.J, Mathiharan, Y.K, Yang, S, Stone, A.C, Wacker, D. | | Deposit date: | 2022-02-11 | | Release date: | 2023-08-16 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Substrate binding and inhibition of the anion exchanger 1 transporter.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7TY8
 
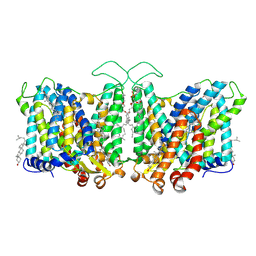 | | Cryo-EM structure of human Anion Exchanger 1 bound to Niflumic Acid | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-{[3-(TRIFLUOROMETHYL)PHENYL]AMINO}NICOTINIC ACID, ... | | Authors: | Capper, M.J, Mathiharan, Y.K, Yang, S, Stone, A.C, Wacker, D. | | Deposit date: | 2022-02-11 | | Release date: | 2023-08-16 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Substrate binding and inhibition of the anion exchanger 1 transporter.
Nat.Struct.Mol.Biol., 30, 2023
|
|
4N0M
 
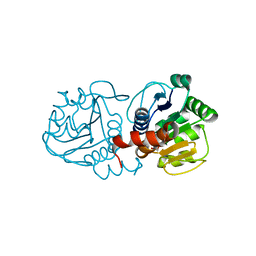 | | Crystal structure of human C53A DJ-1 in complex with Cu | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, Protein DJ-1 | | Authors: | Cendron, L, Girotto, S, Bisaglia, M, Tessari, I, Mammi, S, Zanotti, G, Bubacco, L. | | Deposit date: | 2013-10-02 | | Release date: | 2014-03-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | DJ-1 Is a Copper Chaperone Acting on SOD1 Activation.
J.Biol.Chem., 289, 2014
|
|
6HSJ
 
 | | Crystal structure of the zebrafish peroxisomal SCP2-thiolase (type-1) in complex with CoA | | Descriptor: | ACETATE ION, COENZYME A, GLYCEROL, ... | | Authors: | Wierenga, R.K, Kiema, T.R, Thapa, C.J. | | Deposit date: | 2018-10-01 | | Release date: | 2019-01-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | The peroxisomal zebrafish SCP2-thiolase (type-1) is a weak transient dimer as revealed by crystal structures and native mass spectrometry.
Biochem. J., 476, 2019
|
|
6X5I
 
 | |
5G08
 
 | | Crystal structure of Drosophila NCS-1 bound to chlorpromazine | | Descriptor: | 1,2-ETHANEDIOL, 3-(2-chloro-10H-phenothiazin-10-yl)-N,N-dimethylpropan-1-amine, CALCIUM ION, ... | | Authors: | Chaves-Sanjuan, A, Infantes, L, Sanchez-Barrena, M.J. | | Deposit date: | 2016-03-17 | | Release date: | 2017-01-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Interference of the complex between NCS-1 and Ric8a with phenothiazines regulates synaptic function and is an approach for fragile X syndrome.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
3LZS
 
 | | Crystal Structure of HIV-1 CRF01_AE Protease in Complex with Darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, ACETATE ION, HIV-1 protease | | Authors: | Schiffer, C.A, Bandaranayake, R.M. | | Deposit date: | 2010-03-01 | | Release date: | 2010-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Effect of Clade-Specific Sequence Polymorphisms on HIV-1 Protease Activity and Inhibitor Resistance Pathways.
J.Virol., 84, 2010
|
|
4MTC
 
 | | Crystal structure of human C53A DJ-1 | | Descriptor: | 1,2-ETHANEDIOL, Protein DJ-1 | | Authors: | Cendron, L, Girotto, S, Bisaglia, M, Tessari, I, Mammi, S, Zanotti, G, Bubacco, L. | | Deposit date: | 2013-09-19 | | Release date: | 2014-03-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | DJ-1 Is a Copper Chaperone Acting on SOD1 Activation.
J.Biol.Chem., 289, 2014
|
|
5IH3
 
 | | Crystal structure of mouse CARM1 in complex with SAH at 1.8 Angstroms resolution | | Descriptor: | 1,2-DIMETHOXYETHANE, 1,2-ETHANEDIOL, 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, ... | | Authors: | Cura, V, Marechal, N, Mailliot, J, Troffer-Charlier, N, Hassenboehler, P, Wurtz, J.M, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2016-02-29 | | Release date: | 2017-03-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of mouse CARM1 in complex with SAH at 1.8 Angstroms resolution
To Be Published
|
|
1B6M
 
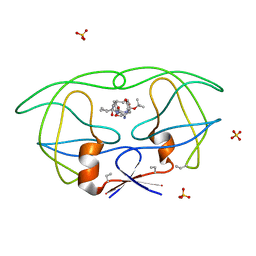 | | HIV-1 PROTEASE COMPLEXED WITH MACROCYCLIC PEPTIDOMIMETIC INHIBITOR 6 | | Descriptor: | RETROPEPSIN, SULFATE ION, [1-BENZYL-3-(8-SEC-BUTYL-7,10-DIOXO-2-OXA-6,9-DIAZA-BICYCLO[11.2.2] HEPTADECA-1(16),13(17),14-TRIEN-11-YLAMINO)-2-HYDROXY-PROPYL]-CARBAMIC ACID TERT-BUTYL ESTER | | Authors: | Martin, J.L, Begun, J, Schindeler, A, Wickramasinghe, W.A, Alewood, D, Alewood, P.F, Bergman, D.A, Brinkworth, R.I, Abbenante, G, March, D.R, Reid, R.C, Fairlie, D.P. | | Deposit date: | 1999-01-17 | | Release date: | 2000-01-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Molecular recognition of macrocyclic peptidomimetic inhibitors by HIV-1 protease.
Biochemistry, 38, 1999
|
|
5IWG
 
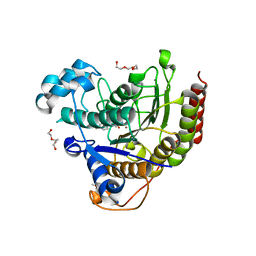 | | HDAC2 WITH LIGAND BRD4884 | | Descriptor: | 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Steinbacher, S. | | Deposit date: | 2016-03-22 | | Release date: | 2016-08-31 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Kinetic and structural insights into the binding of histone deacetylase 1 and 2 (HDAC1, 2) inhibitors.
Bioorg.Med.Chem., 24, 2016
|
|
3MWS
 
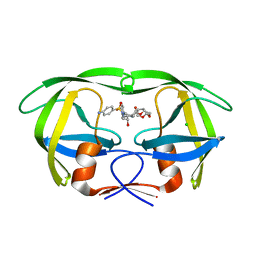 | | Crystal Structure of Group N HIV-1 Protease | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, CHLORIDE ION, HIV-1 Protease | | Authors: | Sayer, J.M, Agniswamy, J, Weber, I.T, Louis, J.M. | | Deposit date: | 2010-05-06 | | Release date: | 2011-03-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Autocatalytic maturation, physical/chemical properties, and crystal structure of group N HIV-1 protease: relevance to drug resistance.
Protein Sci., 19, 2010
|
|
3N3I
 
 | | Crystal Structure of G48V/C95F tethered HIV-1 Protease/Saquinavir complex | | Descriptor: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, Protease | | Authors: | Prashar, V, Bihani, S.C, Das, A, Rao, D.R, Hosur, M.V. | | Deposit date: | 2010-05-20 | | Release date: | 2010-06-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Insights into the mechanism of drug resistance: X-ray structure analysis of G48V/C95F tethered HIV-1 protease dimer/saquinavir complex
Biochem.Biophys.Res.Commun., 396, 2010
|
|
5JCU
 
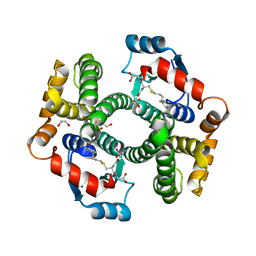 | |
7CUY
 
 | | Crystal structure of Primo-1 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Low molecular weight phosphotyrosine protein phosphatase 1 | | Authors: | Lee, H.S, Kim, S.J, Ku, B. | | Deposit date: | 2020-08-25 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.081 Å) | | Cite: | Structural and Biochemical Characterization of the Two Drosophila Low Molecular Weight-Protein Tyrosine Phosphatases DARP and Primo-1.
Mol.Cells, 43, 2020
|
|
