7PJM
 
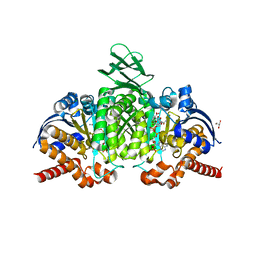 | | Crystal Structure of Ivosidenib-resistant IDH1 variant R132C S280F in complex with NADPH and Ca2+/2-Oxoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Reinbold, R, Rabe, P, Abboud, M.I, Schofield, C.J. | | Deposit date: | 2021-08-24 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Resistance to the isocitrate dehydrogenase 1 mutant inhibitor ivosidenib can be overcome by alternative dimer-interface binding inhibitors.
Nat Commun, 13, 2022
|
|
5EA5
 
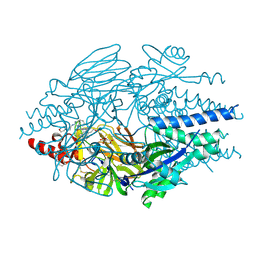 | | Crystal Structure of Inhibitor TMC-353121 in Complex with Prefusion RSV F Glycoprotein | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, 2-[[6-[[[2-(3-hydroxypropyl)-5-methylphenyl]amino]methyl]-2-[[3-(4-morpholinyl)propyl]amino]-1H-benzimidazol-1-yl]methyl]-6-methyl-3-pyridinol, D(-)-TARTARIC ACID, ... | | Authors: | Battles, M.B, McLellan, J.S, Arnoult, E, Roymans, D, Langedijk, J.P. | | Deposit date: | 2015-10-15 | | Release date: | 2015-12-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Molecular mechanism of respiratory syncytial virus fusion inhibitors.
Nat.Chem.Biol., 12, 2016
|
|
7YM7
 
 | |
8P9R
 
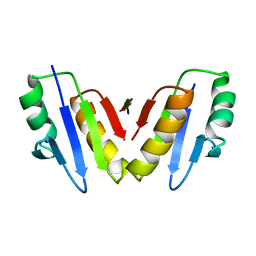 | | Structure of the periplasmic domain of ExbD from E. coli in complex with TonB | | Descriptor: | Biopolymer transport protein ExbD, Protein TonB | | Authors: | Zinke, M, Mechaly, A, Lejeune, M, Haouz, A, Izadi-Pruneyre, N. | | Deposit date: | 2023-06-06 | | Release date: | 2023-09-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Ton motor conformational switch and peptidoglycan role in bacterial nutrient uptake.
Nat Commun, 15, 2024
|
|
7N3V
 
 | | Crystal structure of Mycobacterium smegmatis LmcA | | Descriptor: | GLYCEROL, LmcA, SULFATE ION | | Authors: | Patel, O, Lucet, I, Panjikar, S. | | Deposit date: | 2021-06-02 | | Release date: | 2022-04-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal structure of the putative cell-wall lipoglycan biosynthesis protein LmcA from Mycobacterium smegmatis.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7PVT
 
 | |
5EAH
 
 | | Saccharomyces cerevisiae CYP51 complexed with the plant pathogen inhibitor Difenoconazole | | Descriptor: | 1-[[(2~{R},4~{R})-2-[2-chloranyl-4-(4-chloranylphenoxy)phenyl]-4-methyl-1,3-dioxolan-2-yl]methyl]-1,2,4-triazole, 1-[[(2~{R},4~{S})-2-[2-chloranyl-4-(4-chloranylphenoxy)phenyl]-4-methyl-1,3-dioxolan-2-yl]methyl]-1,2,4-triazole, 1-[[(2~{S},4~{R})-2-[2-chloranyl-4-(4-chloranylphenoxy)phenyl]-4-methyl-1,3-dioxolan-2-yl]methyl]-1,2,4-triazole, ... | | Authors: | Tyndall, J.D.A, Sabherwal, M, Keniya, M.V, Wilson, R.K, Woods, M.V, Monk, B.C. | | Deposit date: | 2015-10-16 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.541 Å) | | Cite: | Structural and Functional Elucidation of Yeast Lanosterol 14 alpha-Demethylase in Complex with Agrochemical Antifungals.
PLoS ONE, 11, 2016
|
|
7N9G
 
 | | Crystal structure of the Abl 1b Kinase domain in complex with Dasatinib and Imatinib | | Descriptor: | 4-(4-METHYL-PIPERAZIN-1-YLMETHYL)-N-[4-METHYL-3-(4-PYRIDIN-3-YL-PYRIMIDIN-2-YLAMINO)-PHENYL]-BENZAMIDE, N-(2-CHLORO-6-METHYLPHENYL)-2-({6-[4-(2-HYDROXYETHYL)PIPERAZIN-1-YL]-2-METHYLPYRIMIDIN-4-YL}AMINO)-1,3-THIAZOLE-5-CARBOXAMIDE, PHOSPHATE ION, ... | | Authors: | Miller, D.J, Xie, T. | | Deposit date: | 2021-06-17 | | Release date: | 2022-04-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Imatinib can act as an Allosteric Activator of Abl Kinase.
J.Mol.Biol., 434, 2022
|
|
7W2C
 
 | | The closed conformation of the sigma-1 receptor from Xenopus laevis complexed with PRE084 | | Descriptor: | 2-morpholin-4-ylethyl 1-phenylcyclohexane-1-carboxylate, GOLD ION, Sigma non-opioid intracellular receptor 1, ... | | Authors: | Meng, F, Sun, Z, Zhou, X. | | Deposit date: | 2021-11-23 | | Release date: | 2022-03-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.335 Å) | | Cite: | An open-like conformation of the sigma-1 receptor reveals its ligand entry pathway.
Nat Commun, 13, 2022
|
|
8SME
 
 | | Structure of SPO1 phage Tad2 in apo state | | Descriptor: | Gp34.65 | | Authors: | Lu, A, Yirmiya, E, Leavitt, A, Avraham, C, Osterman, I, Garb, J, Antine, S.P, Mooney, S.E, Hobbs, S.J, Amitai, G, Sorek, R, Kranzusch, P.J. | | Deposit date: | 2023-04-26 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Phages overcome bacterial immunity via diverse anti-defence proteins.
Nature, 625, 2024
|
|
7N5M
 
 | | PCNA from Thermococcus gammatolerans: crystal III, collection 1, 2.00 A, 1.91 MGy | | Descriptor: | DNA polymerase sliding clamp, GLYCEROL, SULFATE ION | | Authors: | Marin-Tovar, Y, Rudino-Pinera, E. | | Deposit date: | 2021-06-05 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | PCNA from Thermococcus gammatolerans: A protein involved in chromosomal DNA metabolism intrinsically resistant at high levels of ionizing radiation.
Proteins, 90, 2022
|
|
5DK8
 
 | |
8T1U
 
 | | Crystal structure of the DRM2-CTA DNA complex | | Descriptor: | DNA (5'-D(P*AP*TP*TP*AP*TP*TP*AP*AP*TP*(C49)P*TP*AP*AP*AP*TP*TP*TP*A)-3'), DNA (5'-D(P*TP*AP*AP*AP*TP*TP*TP*AP*GP*AP*TP*TP*AP*AP*TP*AP*AP*T)-3'), DNA (cytosine-5)-methyltransferase DRM2, ... | | Authors: | Chen, J, Lu, J, Song, J. | | Deposit date: | 2023-06-03 | | Release date: | 2023-11-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | DNA conformational dynamics in the context-dependent non-CG CHH methylation by plant methyltransferase DRM2.
J.Biol.Chem., 299, 2023
|
|
5HMZ
 
 | | Dengue serotype 3 RNA-dependent RNA polymerase bound to compound 23 | | Descriptor: | 5-(5-(3-hydroxyprop-1-yn-1-yl)thiophen-2-yl)-4-methoxy-2-methyl-N-(methylsulfonyl)benzamide, RNA-directed RNA polymerase NS5, ZINC ION | | Authors: | Noble, C.G. | | Deposit date: | 2016-01-17 | | Release date: | 2016-03-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Discovery of Potent Non-Nucleoside Inhibitors of Dengue Viral RNA-Dependent RNA Polymerase from a Fragment Hit Using Structure-Based Drug Design
J.Med.Chem., 59, 2016
|
|
5DKD
 
 | | Crystal structure of the bromodomain of human BRG1 (SMARCA4) in complex with PFI-3 chemical probe | | Descriptor: | (2E)-1-(2-hydroxyphenyl)-3-[(1R,4R)-5-(pyridin-2-yl)-2,5-diazabicyclo[2.2.1]hept-2-yl]prop-2-en-1-one, 1,2-ETHANEDIOL, Transcription activator BRG1, ... | | Authors: | Tallant, C, Owen, D.R, Gerstenberger, B.S, Fedorov, O, Savitsky, P, Nunez-Alonso, G, Newman, J.A, Filippakopoulos, P, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Muller, S, Knapp, S. | | Deposit date: | 2015-09-03 | | Release date: | 2015-10-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the bromodomain of human BRG1 (SMARCA4) in complex with PFI-3 chemical probe
To Be Published
|
|
7PW0
 
 | |
8VL8
 
 | |
7WAA
 
 | | Crystal structure of MCR-1-S treated by AgNO3 | | Descriptor: | Probable phosphatidylethanolamine transferase Mcr-1, SILVER ION | | Authors: | Zhang, Q, Wang, M, Sun, H. | | Deposit date: | 2021-12-13 | | Release date: | 2022-03-16 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Re-sensitization of mcr carrying multidrug resistant bacteria to colistin by silver.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8SMH
 
 | |
8SP1
 
 | |
7N5K
 
 | | PCNA from Thermococcus gammatolerans: crystal II, collection 1, 1.98 A, 3.84 MGy | | Descriptor: | DNA polymerase sliding clamp, GLYCEROL, SULFATE ION | | Authors: | Marin-Tovar, Y, Rudino-Pinera, E. | | Deposit date: | 2021-06-05 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | PCNA from Thermococcus gammatolerans: A protein involved in chromosomal DNA metabolism intrinsically resistant at high levels of ionizing radiation.
Proteins, 90, 2022
|
|
7W2H
 
 | |
5HNC
 
 | | Synchrotron X-ray single crystal diffraction from protein microcrystals via magnetically oriented microcrystal arrays in gels | | Descriptor: | Lysozyme C | | Authors: | Tsukui, S, Kimura, F, Kusaka, K, Baba, S, Mizuno, N, Kimura, T. | | Deposit date: | 2016-01-18 | | Release date: | 2016-07-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Neutron and X-ray single-crystal diffraction from protein microcrystals via magnetically oriented microcrystal arrays in gels.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
6NEL
 
 | | 4-(2-(4-fluorophenyl)-5-hydroxy-6-oxo-1,6-dihydropyridin-3-yl)benzoic acid bound to influenza 2009 pH1N1 endonuclease | | Descriptor: | 4-[2-(4-fluorophenyl)-5-hydroxy-6-oxo-1,6-dihydropyridin-3-yl]benzoic acid, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Bauman, J.D, Arnold, E. | | Deposit date: | 2018-12-17 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Aryl and Arylalkyl Substituted 3-Hydroxypyridin-2(1H)-ones: Synthesis and Evaluation as Inhibitors of Influenza A Endonuclease.
Chemmedchem, 14, 2019
|
|
7PVW
 
 | |
