6P3Q
 
 | | Calpain-5 (CAPN5) Protease Core (PC) | | Descriptor: | Calpain-5 | | Authors: | Velez, G, Sun, Y.J, Khan, S, Yang, J, Koster, H.J, Lokesh, G, Mahajan, V. | | Deposit date: | 2019-05-24 | | Release date: | 2020-02-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Insights into the Unique Activation Mechanisms of a Non-classical Calpain and Its Disease-Causing Variants.
Cell Rep, 30, 2020
|
|
6P63
 
 | |
6P77
 
 | | 2.5 Angstrom structure of Caci_6494 from Catenulispora Acidiphila | | Descriptor: | Aromatic-ring-hydroxylating dioxygenase beta subunit, GLYCEROL, TETRAETHYLENE GLYCOL, ... | | Authors: | Vuksanovic, N, Silvaggi, N.R. | | Deposit date: | 2019-06-05 | | Release date: | 2020-06-17 | | Last modified: | 2020-09-02 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Structural characterization of three noncanonical NTF2-like superfamily proteins: implications for polyketide biosynthesis.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
3IX9
 
 | |
4IOY
 
 | |
4IO0
 
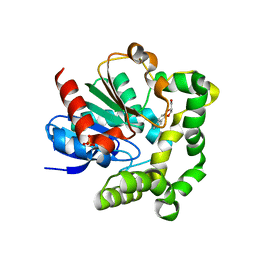 | | Crystal structure of F128A mutant of an epoxide hydrolase from Bacillus megaterium complexed with its product (R)-3-[1]naphthyloxy-propane-1,2-diol | | Descriptor: | (2R)-3-(naphthalen-1-yloxy)propane-1,2-diol, SULFATE ION, Soluble epoxide hydrolase | | Authors: | Kong, X.D, Zhou, J.H, Xu, J.H. | | Deposit date: | 2013-01-07 | | Release date: | 2014-02-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Engineering of an epoxide hydrolase for efficient bioresolution of bulky pharmaco substrates.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
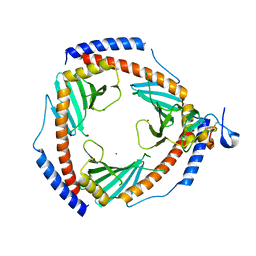
6SZW
 
 | |
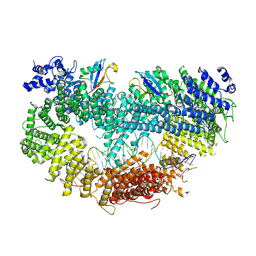
6VAE
 
 | |
6V2J
 
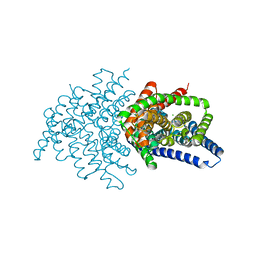 | | Crystal structure of ClC-ec1 triple mutant (E113Q, E148Q, E203Q) | | Descriptor: | CHLORIDE ION, H(+)/Cl(-) exchange transporter ClcA | | Authors: | Maduke, M, Mathews, I.I, Chavan, T.S. | | Deposit date: | 2019-11-24 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | A CLC-ec1 mutant reveals global conformational change and suggests a unifying mechanism for the CLC Cl - /H + transport cycle.
Elife, 9, 2020
|
|
6VDK
 
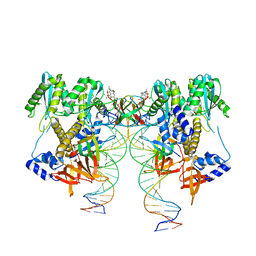 | | CryoEM structure of HIV-1 conserved Intasome Core | | Descriptor: | (4R,12aS)-N-(2,4-difluorobenzyl)-7-hydroxy-4-methyl-6,8-dioxo-3,4,6,8,12,12a-hexahydro-2H-pyrido[1',2':4,5]pyrazino[2,1-b][1,3]oxazine-9-carboxamide, DNA (25-MER), DNA (27-MER), ... | | Authors: | Li, M, Chen, X, Craigie, R. | | Deposit date: | 2019-12-27 | | Release date: | 2020-02-05 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | A Peptide Derived from Lens Epithelium-Derived Growth Factor Stimulates HIV-1 DNA Integration and Facilitates Intasome Structural Studies.
J.Mol.Biol., 432, 2020
|
|
6SUA
 
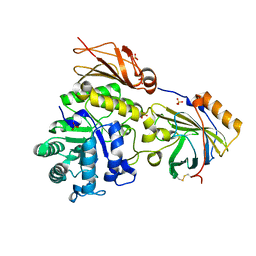 | |
6T13
 
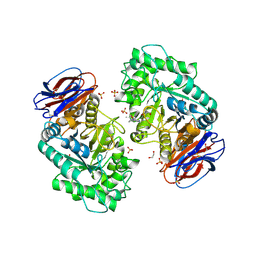 | | CRYSTAL STRUCTURE OF GLUCOCEREBROSIDASE IN COMPLEX WITH A PYRROLOPYRAZINE | | Descriptor: | 1,2-ETHANEDIOL, 1-[4-[2-(4-methoxyphenyl)-5-methyl-pyrrolo[2,3-b]pyrazin-6-yl]piperidin-1-yl]ethanone, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Benz, J, Ehler, A, Hug, M, Huber, S, Rufer, A.C, Guba, W, Jagasia, R, Hofmann, E.C, Rodriguez Sarmiento, R.M. | | Deposit date: | 2019-10-03 | | Release date: | 2020-12-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Novel beta-Glucocerebrosidase Activators That Bind to a New Pocket at a Dimer Interface and Induce Dimerization.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
4IE1
 
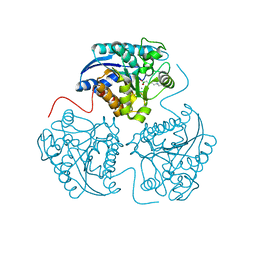 | | Crystal structure of human Arginase-1 complexed with inhibitor 1h | | Descriptor: | Arginase-1, MANGANESE (II) ION, [(5R)-5-amino-5-carboxy-8-hydroxyoctyl](trihydroxy)borate(1-) | | Authors: | Cousido-Siah, A, Mitschler, A, Ruiz, F.X, Beckett, P, Van Zandt, M.C, Ji, M.K, Whitehouse, D, Ryder, T, Jagdmann, E, Andreoli, M, Mazur, A, Padmanilayam, M, Schroeter, H, Golebiowski, A, Podjarny, A. | | Deposit date: | 2012-12-13 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.0006 Å) | | Cite: | 2-Substituted-2-amino-6-boronohexanoic acids as arginase inhibitors.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
6SUN
 
 | | Amicoumacin kinase hAmiN in complex with AMP-PNP, Ca2+ and Ami | | Descriptor: | APH domain-containing protein, amicoumacin kinase, Amicoumacin A, ... | | Authors: | Bourenkov, G.P, Mokrushina, Y.A, Terekhov, S.S, Smirnov, I.V, Gabibov, A.G, Altman, S. | | Deposit date: | 2019-09-16 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | A kinase bioscavenger provides antibiotic resistance by extremely tight substrate binding.
Sci Adv, 6, 2020
|
|
6SJ0
 
 | | Amidohydrolase, AHS | | Descriptor: | Amidohydrolase, BICARBONATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Naismith, J.H, Song, H. | | Deposit date: | 2019-08-12 | | Release date: | 2020-01-15 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The Biosynthesis of the Benzoxazole in Nataxazole Proceeds via an Unstable Ester and has Synthetic Utility.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6SME
 
 | |
6VAN
 
 | | Crystal structure of caltubin from the great pond snail | | Descriptor: | 1,2-ETHANEDIOL, Caltubin, EF-hand, ... | | Authors: | Dong, A, Li, A, Zhang, Q, Barszczyk, A, Chern, Y.H, Arrowsmith, C.H, Edwards, A.M, Zhong, Z.P, Tong, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-12-17 | | Release date: | 2020-12-23 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Cell-penetrating caltubin promotes neurite outgrowth and regrowth through calcium-dependent microtubule regulation
to be published
|
|
4IK4
 
 | | High resolution structure of GCaMP3 at pH 5.0 | | Descriptor: | CALCIUM ION, RCaMP, Green fluorescent protein | | Authors: | Chen, Y, Song, X, Miao, L, Zhu, Y, Ji, G. | | Deposit date: | 2012-12-25 | | Release date: | 2014-02-05 | | Last modified: | 2017-06-21 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural insight into enhanced calcium indicator GCaMP3 and GCaMPJ to promote further improvement.
Protein Cell, 4, 2013
|
|
6PDT
 
 | | cryoEM structure of yeast glucokinase filament | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Glucokinase-1, MAGNESIUM ION, ... | | Authors: | Lynch, E.M, Dosey, A.M, Farrell, D.P, Stoddard, P.R, Kollman, J.M. | | Deposit date: | 2019-06-19 | | Release date: | 2020-03-11 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Polymerization in the actin ATPase clan regulates hexokinase activity in yeast.
Science, 367, 2020
|
|
4INZ
 
 | | The crystal structure of M145A mutant of an epoxide hydrolase from Bacillus megaterium | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Soluble epoxide hydrolase | | Authors: | Kong, X.D, Zhou, J.H, Xu, J.H. | | Deposit date: | 2013-01-07 | | Release date: | 2014-02-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Engineering of an epoxide hydrolase for efficient bioresolution of bulky pharmaco substrates.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
6SIX
 
 | | PaaK family AMP-ligase with ANP | | Descriptor: | 1,2-ETHANEDIOL, AMP-dependent synthetase and ligase, CITRATE ANION, ... | | Authors: | Naismith, J.H, Song, H. | | Deposit date: | 2019-08-12 | | Release date: | 2020-01-15 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | The Biosynthesis of the Benzoxazole in Nataxazole Proceeds via an Unstable Ester and has Synthetic Utility.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6VCH
 
 | | Crystal structure of Nitrosotalea devanaterra carotenoid cleavage dioxygenase in complex with 3-hydroxy-beta-apo-14'-carotenal | | Descriptor: | (2E,4E,6E,8E,10E)-11-[(4R)-4-hydroxy-2,6,6-trimethylcyclohex-1-en-1-yl]-5,9-dimethylundeca-2,4,6,8,10-pentaenal, CHLORIDE ION, COBALT (II) ION, ... | | Authors: | Daruwalla, A, Shi, W, Kiser, P.D. | | Deposit date: | 2019-12-20 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis for carotenoid cleavage by an archaeal carotenoid dioxygenase.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6VEA
 
 | | Structure of the Glutamate-Like Receptor GLR3.2 ligand-binding domain in complex with Glycine | | Descriptor: | BETA-MERCAPTOETHANOL, GLYCINE, Glutamate receptor 3.2, ... | | Authors: | Gangwar, S.P, Green, M.N, Yoder, J.B, Sobolevsky, A.I. | | Deposit date: | 2019-12-30 | | Release date: | 2020-09-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structure of the Arabidopsis Glutamate Receptor-like Channel GLR3.2 Ligand-Binding Domain.
Structure, 29, 2021
|
|
6VE8
 
 | | Structure of the Glutamate-Like Receptor GLR3.2 ligand-binding domain in complex with Methionine | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Gangwar, S.P, Green, M.N, Yoder, J.B, Sobolevsky, A.I. | | Deposit date: | 2019-12-30 | | Release date: | 2020-09-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of the Arabidopsis Glutamate Receptor-like Channel GLR3.2 Ligand-Binding Domain.
Structure, 29, 2021
|
|
6PIF
 
 | | V. cholerae TniQ-Cascade complex, open conformation | | Descriptor: | Cas7, type I-F CRISPR-associated protein, TniQ monomer 1, ... | | Authors: | Halpin-Healy, T, Klompe, S, Sternberg, S.H. | | Deposit date: | 2019-06-26 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of DNA targeting by a transposon-encoded CRISPR-Cas system.
Nature, 577, 2020
|
|
