1TKX
 
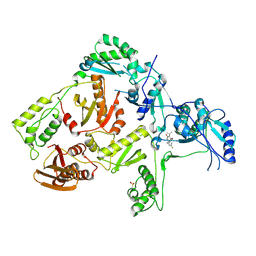 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH GW490745 | | Descriptor: | 4-[(CYCLOPROPYLETHYNYL)OXY]-6-FLUORO-3-ISOPROPYLQUINOLIN-2(1H)-ONE, Pol polyprotein, Reverse transcriptase, ... | | Authors: | Ren, J, Hopkins, A.L, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2004-06-09 | | Release date: | 2004-12-07 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Design of non-nucleoside inhibitors of HIV-1 reverse transcriptase with improved drug resistance properties. 2.
J.Med.Chem., 47, 2004
|
|
1TKZ
 
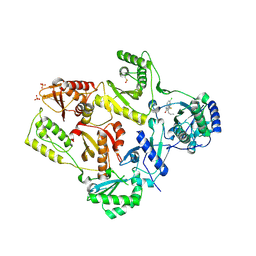 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH GW429576 | | Descriptor: | 6-CHLORO-4-(CYCLOHEXYLSULFANYL)-3-PROPYLQUINOLIN-2(1H)-ONE, PHOSPHATE ION, Pol polyprotein, ... | | Authors: | Hopkins, A.L, Ren, J, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2004-06-09 | | Release date: | 2004-12-07 | | Last modified: | 2020-01-15 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Design of non-nucleoside inhibitors of HIV-1 reverse transcriptase with improved drug resistance properties. 1.
J.Med.Chem., 47, 2004
|
|
7ZR0
 
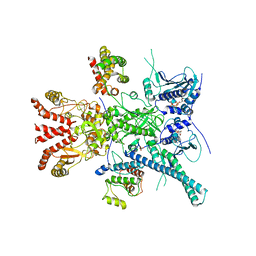 | | CryoEM structure of HSP90-CDC37-BRAF(V600E) complex. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Heat shock protein HSP 90-beta, Hsp90 co-chaperone Cdc37, ... | | Authors: | Oberoi, J, Pearl, L.H. | | Deposit date: | 2022-05-03 | | Release date: | 2022-12-14 | | Last modified: | 2023-01-11 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | HSP90-CDC37-PP5 forms a structural platform for kinase dephosphorylation.
Nat Commun, 13, 2022
|
|
7ZR5
 
 | |
7ZR6
 
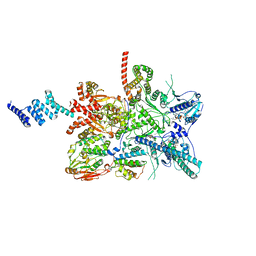 | |
1FMB
 
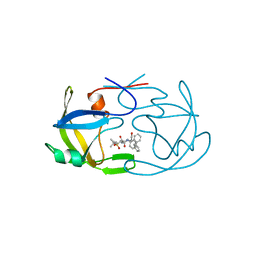 | | EIAV PROTEASE COMPLEXED WITH THE INHIBITOR HBY-793 | | Descriptor: | EIAV PROTEASE, [2-(2-METHYL-PROPANE-2-SULFONYLMETHYL)-3-NAPHTHALEN-1-YL-PROPIONYL-VALINYL]-PHENYLALANINOL | | Authors: | Wlodawer, A, Gustchina, A, Zdanov, A, Kervinen, J. | | Deposit date: | 1996-02-27 | | Release date: | 1996-10-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of equine infectious anemia virus proteinase complexed with an inhibitor.
Protein Sci., 5, 1996
|
|
1PRO
 
 | | HIV-1 PROTEASE DIMER COMPLEXED WITH A-98881 | | Descriptor: | (5R,6R)-2,4-BIS-(4-HYDROXY-3-METHOXYBENZYL)-1,5-DIBENZYL-3-OXO-6-HYDROXY-1,2,4-TRIAZACYCLOHEPTANE, HIV-1 PROTEASE | | Authors: | Park, C.H, Kong, X.P, Dealwis, C.G. | | Deposit date: | 1995-07-18 | | Release date: | 1996-08-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A novel, picomolar inhibitor of human immunodeficiency virus type 1 protease.
J.Med.Chem., 39, 1996
|
|
6LUR
 
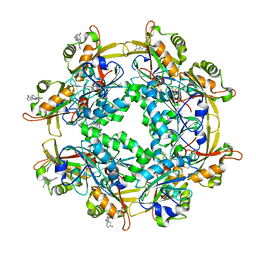 | |
8FFW
 
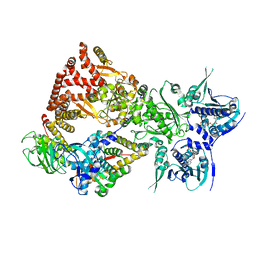 | | Cryo-EM structure of the GR-Hsp90-FKBP51 complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DEXAMETHASONE, Glucocorticoid receptor, ... | | Authors: | Noddings, C.M, Agard, D.A. | | Deposit date: | 2022-12-10 | | Release date: | 2023-11-01 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Cryo-EM reveals how Hsp90 and FKBP immunophilins co-regulate the glucocorticoid receptor.
Nat.Struct.Mol.Biol., 30, 2023
|
|
5EAW
 
 | | Crystal structure of Dna2 nuclease-helicase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA replication ATP-dependent helicase/nuclease DNA2, IRON/SULFUR CLUSTER | | Authors: | Zhou, C, Pourmal, S, Pavletich, N.P. | | Deposit date: | 2015-10-17 | | Release date: | 2015-11-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Dna2 nuclease-helicase structure, mechanism and regulation by Rpa.
Elife, 4, 2015
|
|
2C3S
 
 | | Structure Of Sars Cov Main Proteinase At 1.9 A (Ph6.5) | | Descriptor: | SARS COV 3C-LIKE PROTEINASE | | Authors: | Xu, T, Ooi, A, Lee, H.-C, Lescar, J. | | Deposit date: | 2005-10-12 | | Release date: | 2005-10-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the Sars Coronavirus Main Proteinase as an Active C2 Crystallographic Dimer.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
3U50
 
 | | Crystal Structure of the Tetrahymena telomerase processivity factor Teb1 OB-C | | Descriptor: | Telomerase-associated protein 82, ZINC ION | | Authors: | Zeng, Z, Huang, J, Yang, Y, Lei, M. | | Deposit date: | 2011-10-10 | | Release date: | 2011-12-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for Tetrahymena telomerase processivity factor Teb1 binding to single-stranded telomeric-repeat DNA.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3U58
 
 | | Crystal Structure of the Tetrahymena telomerase processivity factor Teb1 AB | | Descriptor: | DNA (5'-D(*GP*GP*GP*T)-3'), Tetrahymena Teb1 AB | | Authors: | Zeng, Z, Huang, J, Yang, Y, Lei, M. | | Deposit date: | 2011-10-11 | | Release date: | 2011-12-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.613 Å) | | Cite: | Structural basis for Tetrahymena telomerase processivity factor Teb1 binding to single-stranded telomeric-repeat DNA.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
8BAO
 
 | |
6CBZ
 
 | |
3U4V
 
 | |
3U4Z
 
 | |
6EY7
 
 | | Human cytomegalovirus terminase nuclease domain, Mn soaked, inhibitor bound | | Descriptor: | 4-[(4-fluorophenyl)methyl-methyl-amino]-2,4-bis(oxidanylidene)butanoic acid, CHLORIDE ION, MANGANESE (II) ION, ... | | Authors: | Bongarzone, S, Nadal, M, Kaczmarska, Z, Machon, C, Alvarez, M, Albericio, F, Coll, M. | | Deposit date: | 2017-11-10 | | Release date: | 2018-10-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure-Driven Discovery of alpha , gamma-Diketoacid Inhibitors Against UL89 Herpesvirus Terminase.
Acs Omega, 3, 2018
|
|
5OKC
 
 | |
8CI4
 
 | |
2J9J
 
 | | Atomic-resolution Crystal Structure of Chemically-Synthesized HIV-1 Protease Complexed with Inhibitor JG-365 | | Descriptor: | ACETATE ION, GLYCEROL, INHIBITOR MOLECULE JG365, ... | | Authors: | Malito, E, Shen, Y, Johnson, E.C.B, Tang, W.J. | | Deposit date: | 2006-11-11 | | Release date: | 2007-08-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Insights from Atomic-Resolution X-Ray Structures of Chemically Synthesized HIV-1 Protease in Complex with Inhibitors.
J.Mol.Biol., 373, 2007
|
|
8DAW
 
 | |
8DAU
 
 | |
8DAR
 
 | |
8DAT
 
 | |
