5LHN
 
 | | The catalytic domain of murine urokinase-type plasminogen activator in complex with the allosteric inhibitory nanobody Nb7 | | Descriptor: | 1,2-ETHANEDIOL, Camelid-Derived Antibody Fragment Nb7, SULFATE ION, ... | | Authors: | Kromann-Hansen, T, Lange, E.L, Sorensen, H.P, Ghassabeh, G.H, Huang, M, Jensen, J.K, Muyldermans, S, Declerck, P, Andreasen, P.A. | | Deposit date: | 2016-07-12 | | Release date: | 2017-06-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Discovery of a novel conformational equilibrium in urokinase-type plasminogen activator.
Sci Rep, 7, 2017
|
|
5LHQ
 
 | | The EGR-cmk active site inhibited catalytic domain of murine urokinase-type plasminogen activator in complex with the allosteric inhibitory nanobody Nb7 | | Descriptor: | 1,2-ETHANEDIOL, Camelid-Derived Antibody Fragment Nb7, L-alpha-glutamyl-N-{(1S)-4-{[amino(iminio)methyl]amino}-1-[(1S)-2-chloro-1-hydroxyethyl]butyl}glycinamide, ... | | Authors: | Kromann-Hansen, T, Lange, E.L, Sorensen, H.P, Ghassabeh, G.H, Huang, M, Jensen, J.K, Muyldermans, S, Declerck, P.J, Andreasen, P.A. | | Deposit date: | 2016-07-12 | | Release date: | 2017-06-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of a novel conformational equilibrium in urokinase-type plasminogen activator.
Sci Rep, 7, 2017
|
|
7KBR
 
 | | Co-crystal structure of alpha glucosidase with compound 10 | | Descriptor: | 1,2-ETHANEDIOL, 2-{[2-nitro-4-(triazan-1-yl)phenyl]amino}ethyl (2-{[(1S,2S,3R,4S,5S)-2,3,4,5-tetrahydroxy-5-(hydroxymethyl)cyclohexyl]amino}ethyl)carbamate, CALCIUM ION, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2020-10-02 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | N-Substituted Valiolamine Derivatives as Potent Inhibitors of Endoplasmic Reticulum alpha-Glucosidases I and II with Antiviral Activity.
J.Med.Chem., 64, 2021
|
|
7L9E
 
 | | Crystal structure of apo-alpha glucosidase | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2021-01-03 | | Release date: | 2021-12-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | N-Substituted Valiolamine Derivatives as Potent Inhibitors of Endoplasmic Reticulum alpha-Glucosidases I and II with Antiviral Activity.
J.Med.Chem., 64, 2021
|
|
6VVU
 
 | | Anti-Tryptase fab E104.v1 bound to tryptase | | Descriptor: | CALCIUM ION, Fab E104.v1 heavy chain, Fab E104.v1 light chain, ... | | Authors: | Ultsch, M, Koerber, J.T. | | Deposit date: | 2020-02-18 | | Release date: | 2020-12-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Bivalent antibody pliers inhibit beta-tryptase by an allosteric mechanism dependent on the IgG hinge.
Nat Commun, 11, 2020
|
|
8B04
 
 | | STRUCTURE OF PORCINE PANCREATIC ELASTASE BOUND TO A FRAGMENT OF AN ISOXAZOLONE INHIBITOR | | Descriptor: | 2-cyclopropylcarbonyl-3-propan-2-yl-1,2-oxazol-5-one, CALCIUM ION, Chymotrypsin-like elastase family member 1, ... | | Authors: | Ferraroni, M, Giovannoni, P, Masini, V. | | Deposit date: | 2022-09-07 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray structural study of human neutrophil elastase inhibition with a series of azaindoles, azaindazoles and isoxazolones
J.Mol.Struct., 1274, 2023
|
|
8B1Y
 
 | | STRUCTURE OF PORCINE PANCREATIC ELASTASE BOUND TO A FRAGMENT OF A 5-AZAINDAZOLE INHIBITOR | | Descriptor: | 1-cyclopropylcarbonylpyrazolo[4,3-c]pyridine-3-carbonitrile, CALCIUM ION, Chymotrypsin-like elastase family member 1, ... | | Authors: | Ferraroni, M, Giovannoni, P, Gerace, A. | | Deposit date: | 2022-09-12 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | X-ray structural study of human neutrophil elastase inhibition with a series of azaindoles, azaindazoles and isoxazolones
J.Mol.Struct., 1274, 2023
|
|
8B49
 
 | | STRUCTURE OF PORCINE PANCREATIC ELASTASE BOUND TO A FRAGMENT (m-toluoylcarbonyl group) OF A 5-AZAINDOLE INHIBITOR | | Descriptor: | 1-(3-methylphenyl)carbonylpyrrolo[3,2-c]pyridine-3-carbonitrile, CALCIUM ION, Chymotrypsin-like elastase family member 1, ... | | Authors: | Ferraroni, M, Giovannoni, P, Gerace, A. | | Deposit date: | 2022-09-20 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | X-ray structural study of human neutrophil elastase inhibition with a series of azaindoles, azaindazoles and isoxazolones
J.Mol.Struct., 1274, 2023
|
|
8B53
 
 | |
6QH9
 
 | | Crystal Structure of Human Kallikrein 6 in complex with GSK3239861A | | Descriptor: | (3~{R})-~{N}-(4-carbamimidoylphenyl)-2-oxidanylidene-piperidine-3-carboxamide, (3~{S})-~{N}-(4-carbamimidoylphenyl)-2-oxidanylidene-piperidine-3-carboxamide, GLYCEROL, ... | | Authors: | Thorpe, J.H. | | Deposit date: | 2019-01-16 | | Release date: | 2019-02-06 | | Last modified: | 2019-02-27 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Kallikrein 5 inhibitors identified through structure based drug design in search for a treatment for Netherton Syndrome.
Bioorg. Med. Chem. Lett., 29, 2019
|
|
6QHB
 
 | | Crystal Structure of Human Kallikrein 6 in complex with GSK578724A | | Descriptor: | GLYCEROL, Kallikrein-6, ~{N}-(4-carbamimidoylphenyl)-3-methoxy-2-oxidanyl-benzamide | | Authors: | Thorpe, J.H. | | Deposit date: | 2019-01-16 | | Release date: | 2019-02-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Kallikrein 5 inhibitors identified through structure based drug design in search for a treatment for Netherton Syndrome.
Bioorg. Med. Chem. Lett., 29, 2019
|
|
6QHA
 
 | | Crystal Structure of Human Kallikrein 6 in complex with GSK3205388B | | Descriptor: | GLYCEROL, Kallikrein-6, UNKNOWN ATOM OR ION, ... | | Authors: | Thorpe, J.H. | | Deposit date: | 2019-01-16 | | Release date: | 2019-02-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Kallikrein 5 inhibitors identified through structure based drug design in search for a treatment for Netherton Syndrome.
Bioorg. Med. Chem. Lett., 29, 2019
|
|
6QHC
 
 | | Crystal Structure of Human Kallikrein 6 in complex with GSK358180B | | Descriptor: | DIMETHYL SULFOXIDE, GLYCEROL, Kallikrein-6, ... | | Authors: | Thorpe, J.H. | | Deposit date: | 2019-01-16 | | Release date: | 2019-02-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Kallikrein 5 inhibitors identified through structure based drug design in search for a treatment for Netherton Syndrome.
Bioorg. Med. Chem. Lett., 29, 2019
|
|
5TO3
 
 | | Crystal structure of thrombin mutant W215A/E217A fused to EGF456 of thrombomodulin via a 31-residue linker and bound to PPACK | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-alpha-D-mannopyranose-(1-4)-[beta-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, ... | | Authors: | Barranco-Medina, S, Murphy, M, Pelc, L, Chen, Z, Di Cera, E, Pozzi, N. | | Deposit date: | 2016-10-16 | | Release date: | 2017-03-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Rational Design of Protein C Activators.
Sci Rep, 7, 2017
|
|
5WI6
 
 | | Human beta-1 tryptase mutant Ile99Cys | | Descriptor: | L-alpha-glutamyl-N-{(1S)-4-{[amino(iminio)methyl]amino}-1-[(1S)-2-chloro-1-hydroxyethyl]butyl}glycinamide, SULFATE ION, Tryptase alpha/beta-1 | | Authors: | Eigenbrot, C, Maun, H.R. | | Deposit date: | 2017-07-18 | | Release date: | 2018-04-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Dual functionality of beta-tryptase protomers as both proteases and cofactors in the active tetramer.
J. Biol. Chem., 293, 2018
|
|
6EST
 
 | |
6Q1U
 
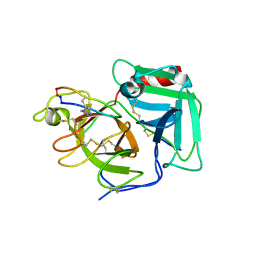 | | Structure of plasmin and peptide complex | | Descriptor: | 1-methyl-1H-1,2,3-triazole, GLY-ARG-ALA-TYR-LYS-SER-LYS-PRO-PRO-ILE-ALA-PHE-PRO-ASP, Plasminogen | | Authors: | Wu, G, Law, R.H.P. | | Deposit date: | 2019-08-06 | | Release date: | 2020-04-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3467 Å) | | Cite: | Application and Structural Analysis of Triazole-Bridged Disulfide Mimetics in Cyclic Peptides.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
4SGA
 
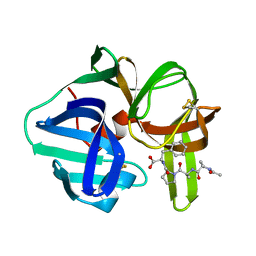 | |
7CBK
 
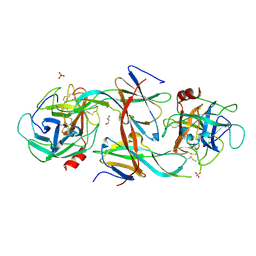 | | Structure of Human Neutrophil Elastase Ecotin complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Ecotin, ... | | Authors: | Jobichen, C, Sivaraman, J. | | Deposit date: | 2020-06-12 | | Release date: | 2020-08-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis for the Inhibition Mechanism of Ecotin against Neutrophil Elastase by Targeting the Active Site and Secondary Binding Site.
Biochemistry, 59, 2020
|
|
7THS
 
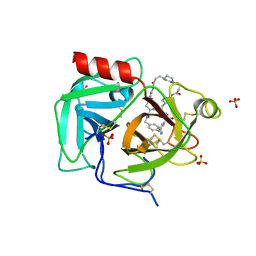 | | Macrocyclic plasmin inhibitor | | Descriptor: | (2S)-butane-1,2-diol, (6S,9R,20R,23S)-N-{[4-(aminomethyl)phenyl]methyl}-20-[(benzenesulfonyl)amino]-3,13,21-trioxo-2,6,9,14,22-pentaazatetracyclo[23.2.2.2~6,9~.2~15,18~]tritriaconta-1(27),15,17,25,28,30-hexaene-23-carboxamide, Plasminogen, ... | | Authors: | Guojie, W. | | Deposit date: | 2022-01-12 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Synthesis and Structural Characterization of Macrocyclic Plasmin Inhibitors.
Chemmedchem, 18, 2023
|
|
7PQO
 
 | | Catalytic fragment of MASP-1 in complex with P1 site mutant ecotin | | Descriptor: | Ecotin, GLYCEROL, Mannan-binding lectin serine protease 1, ... | | Authors: | Harmat, V, Fodor, K, Heja, D. | | Deposit date: | 2021-09-17 | | Release date: | 2022-05-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.39 Å) | | Cite: | Synergy of protease-binding sites within the ecotin homodimer is crucial for inhibition of MASP enzymes and for blocking lectin pathway activation.
J.Biol.Chem., 298, 2022
|
|
7UAH
 
 | | Macrocyclic plasmin inhibitor | | Descriptor: | (2~{R})-butane-1,2-diol, (6S,9R,19S,22R)-N-{[4-(aminomethyl)phenyl]methyl}-22-[(3-chlorobenzene-1-sulfonyl)amino]-3,12,21-trioxo-2,6,9,13,20-pentaazatetracyclo[22.2.2.2~6,9~.2~14,17~]dotriaconta-1(26),14,16,24,27,29-hexaene-19-carboxamide, Plasminogen, ... | | Authors: | Guojie, W. | | Deposit date: | 2022-03-12 | | Release date: | 2023-03-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Synthesis and Structural Characterization of Macrocyclic Plasmin Inhibitors.
Chemmedchem, 18, 2023
|
|
7ARX
 
 | | Crystal structure of the catalytic fragment of masp-1 in complex with SFMI1 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Mannan-binding lectin serine protease 1, SFMI1 - Sunflower MASP1 inhibitor | | Authors: | Durvanger, Z, Harmat, V, Dobo, J, Megyeri, M. | | Deposit date: | 2020-10-26 | | Release date: | 2021-11-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Directed Evolution-Driven Increase of Structural Plasticity Is a Prerequisite for Binding the Complement Lectin Pathway Blocking MASP-Inhibitor Peptides.
Acs Chem.Biol., 17, 2022
|
|
6Q8S
 
 | |
6QEO
 
 | | Crystal structure of Porcine Pancreatic Elastase (PPE) in complex with the 3-Oxo-beta-Sultam inhibitor LMC269 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-methyl-1-[[1-[(4-nitrophenyl)methyl]-1,2,3-triazol-4-yl]methylamino]-1-oxidanylidene-propane-2-sulfonic acid, Chymotrypsin-like elastase family member 1, ... | | Authors: | Brito, J.A, Almeida, V.T, Carvalho, L.M, Moreira, R, Archer, M. | | Deposit date: | 2019-01-08 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | 3-Oxo-beta-sultam as a Sulfonylating Chemotype for Inhibition of Serine Hydrolases and Activity-Based Protein Profiling.
Acs Chem.Biol., 15, 2020
|
|
