2Y81
 
 | | Structure and property based design of factor Xa inhibitors: pyrrolidin-2-ones with aminoindane and phenylpyrrolidine P4 motifs | | Descriptor: | 6-CHLORO-N-((3S)-2-OXO-1-{4-[(2R)-2--PYRROLIDINYL] PHENYL}-3-PYRROLIDINYL)-2-NAPHTHALENESULFONAMIDE, ACTIVATED FACTOR XA HEAVY CHAIN, CALCIUM ION, ... | | Authors: | Young, R.J, Adams, C, Blows, M, Brown, D, Burns-Kurtis, C.L, Chaudry, L, Chan, C, Convery, M.A, Davies, D.E, Exall, A.M, Foster, G, Harling, J.D, Hortense, E, Irving, W.R, Irvine, S, Jackson, S, Kleanthous, S, Pateman, A.J, Patikis, A.N, Roethka, T.J, Senger, S, Stelman, G.J, Toomey, J.R, West, R.I, Whittaker, C, Zhou, P, Watson, N.S. | | Deposit date: | 2011-02-02 | | Release date: | 2011-03-16 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and Property Based Design of Factor Xa Inhibitors: Pyrrolidin-2-Ones with Aminoindane and Phenylpyrrolidine P4 Motifs.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
2Y80
 
 | | Structure and property based design of factor Xa inhibitors: pyrrolidin-2-ones with aminoindane and phenylpyrrolidine P4 motifs | | Descriptor: | 6-CHLORO-N-{(3S)-1-[(1S)-1-(DIMETHYLAMINO)-2,3-DIHYDRO-1H-INDEN-5-YL]-2-OXO-3-PYRROLIDINYL}-2-NAPHTHALENESULFONAMIDE, ACTIVATED FACTOR XA HEAVY CHAIN, CALCIUM ION, ... | | Authors: | Young, R.J, Adams, C, Blows, M, Brown, D, Burns-Kurtis, C.L, Chaudry, L, Chan, C, Convery, M.A, Davies, D.E, Exall, A.M, Foster, G, Harling, J.D, Hortense, E, Irving, W.R, Irvine, S, Jackson, S, Kleanthous, S, Pateman, A.J, Patikis, A.N, Roethka, T.J, Senger, S, Stelman, G.J, Toomey, J.R, West, R.I, Whittaker, C, Zhou, P, Watson, N.S. | | Deposit date: | 2011-02-02 | | Release date: | 2011-03-16 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and Property Based Design of Factor Xa Inhibitors: Pyrrolidin-2-Ones with Aminoindane and Phenylpyrrolidine P4 Motifs.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
7YSJ
 
 | |
3H5N
 
 | | Crystal structure of E. coli MccB + ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, MccB protein, ... | | Authors: | Regni, C.A, Roush, R.F, Miller, D, Nourse, A, Walsh, C.T, Schulman, B.A. | | Deposit date: | 2009-04-22 | | Release date: | 2009-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | How the MccB bacterial ancestor of ubiquitin E1 initiates biosynthesis of the microcin C7 antibiotic.
Embo J., 28, 2009
|
|
8A8K
 
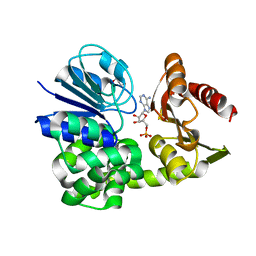 | |
4JHW
 
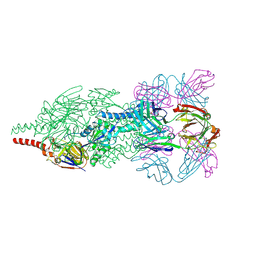 | | Crystal Structure of Respiratory Syncytial Virus Fusion Glycoprotein Stabilized in the Prefusion Conformation by Human Antibody D25 | | Descriptor: | D25 antigen-binding fragment heavy chain, D25 light chain, Fusion glycoprotein F0 | | Authors: | Mclellan, J.S, Chen, M, Leung, S, Graepel, K.W, Du, X, Yang, Y, Zhou, T, Baxa, U, Yasuda, E, Beaumont, T, Kumar, A, Modjarrad, K, Zheng, Z, Zhao, M, Xia, N, Kwong, P.D, Graham, B.S. | | Deposit date: | 2013-03-05 | | Release date: | 2013-05-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structure of RSV fusion glycoprotein trimer bound to a prefusion-specific neutralizing antibody.
Science, 340, 2013
|
|
8B0H
 
 | | 2C9, C5b9-CD59 cryoEM structure | | Descriptor: | CD59 glycoprotein, Complement C5, Complement component C6, ... | | Authors: | Couves, E.C, Gardner, S, Bubeck, D. | | Deposit date: | 2022-09-07 | | Release date: | 2023-02-22 | | Last modified: | 2023-03-01 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for membrane attack complex inhibition by CD59.
Nat Commun, 14, 2023
|
|
8AW0
 
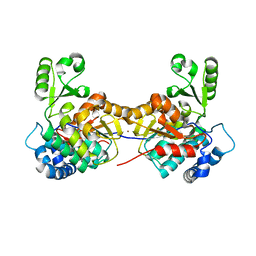 | |
8AVZ
 
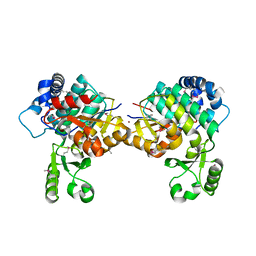 | |
3I4Q
 
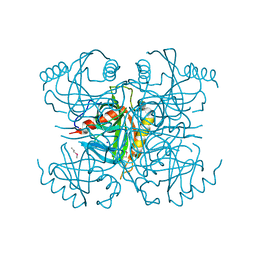 | | Structure of a putative inorganic pyrophosphatase from the oil-degrading bacterium Oleispira antarctica | | Descriptor: | APC40078, SODIUM ION | | Authors: | Singer, A.U, Evdokimova, E, Kagan, O, Edwards, A.M, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-07-02 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Genome sequence and functional genomic analysis of the oil-degrading bacterium Oleispira antarctica.
Nat Commun, 4, 2013
|
|
3HQU
 
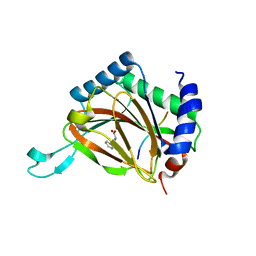 | | PHD2:Fe:UN9:partial HIF1-alpha substrate complex | | Descriptor: | Egl nine homolog 1, FE (II) ION, Hypoxia-inducible factor 1 alpha, ... | | Authors: | Chowdhury, R, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2009-06-08 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for binding of hypoxia-inducible factor to the oxygen-sensing prolyl hydroxylases
Structure, 17, 2009
|
|
4KTY
 
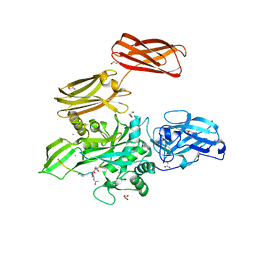 | | Fibrin-stabilizing factor with a bound ligand | | Descriptor: | CALCIUM ION, Coagulation factor XIII A chain, GLYCEROL, ... | | Authors: | Stieler, M, Heine, A, Klebe, G. | | Deposit date: | 2013-05-21 | | Release date: | 2013-12-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure of Active Coagulation Factor XIII Triggered by Calcium Binding: Basis for the Design of Next-Generation Anticoagulants.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
8AQQ
 
 | |
8AQR
 
 | |
3HPB
 
 | |
3HQR
 
 | | PHD2:Mn:NOG:HIF1-alpha substrate complex | | Descriptor: | Egl nine homolog 1, Hypoxia-inducible factor 1 alpha, MANGANESE (II) ION, ... | | Authors: | Chowdhury, R, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2009-06-08 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for binding of hypoxia-inducible factor to the oxygen-sensing prolyl hydroxylases
Structure, 17, 2009
|
|
3H5X
 
 | | Crystal Structure of 2'-amino-2'-deoxy-cytidine-5'-triphosphate bound to Norovirus GII RNA polymerase | | Descriptor: | 2'-amino-2'-deoxycytidine 5'-(tetrahydrogen triphosphate), 5'-R(*UP*GP*CP*CP*CP*GP*GP*G)-3', 5'-R(P*UP*GP*CP*CP*CP*GP*GP*GP*C)-3', ... | | Authors: | Zamyatkin, D.F, Parra, F, Machin, A, Grochulski, P, Ng, K.K.S. | | Deposit date: | 2009-04-22 | | Release date: | 2009-05-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Binding of 2'-amino-2'-deoxycytidine-5'-triphosphate to norovirus polymerase induces rearrangement of the active site.
J.Mol.Biol., 390, 2009
|
|
3H9Q
 
 | | Crystal structure of E. coli MccB + SeMet MccA | | Descriptor: | MccB protein, Microcin C7 ANALOG, SULFATE ION, ... | | Authors: | Regni, C.A, Roush, R.F, Miller, D, Nourse, A, Walsh, C.T, Schulman, B.A. | | Deposit date: | 2009-04-30 | | Release date: | 2009-06-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | How the MccB bacterial ancestor of ubiquitin E1 initiates biosynthesis of the microcin C7 antibiotic.
Embo J., 28, 2009
|
|
3H9J
 
 | | Crystal structure of E. coli MccB + AMPCPP + SeMeT MccA | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, MccB protein, Microcin C7 ANALOG, ... | | Authors: | Regni, C.A, Roush, R.F, Miller, D, Nourse, A, Walsh, C.T, Schulman, B.A. | | Deposit date: | 2009-04-30 | | Release date: | 2009-06-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | How the MccB bacterial ancestor of ubiquitin E1 initiates biosynthesis of the microcin C7 antibiotic.
Embo J., 28, 2009
|
|
4LK4
 
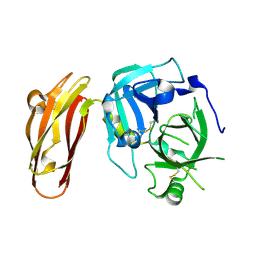 | |
3IGM
 
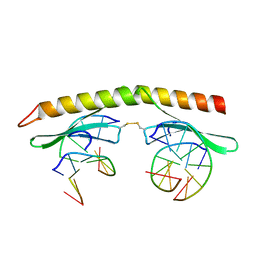 | | A 2.2A crystal structure of the AP2 domain of PF14_0633 from P. falciparum, bound as a domain-swapped dimer to its cognate DNA | | Descriptor: | 5'-D(*TP*GP*CP*AP*TP*GP*CP*A)-3', PF14_0633 protein | | Authors: | Lindner, S.E, De Silva, E, Keck, J.L, Llinas, M. | | Deposit date: | 2009-07-28 | | Release date: | 2009-11-10 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Determinants of DNA Binding by a P. falciparum ApiAP2 Transcriptional Regulator.
J.Mol.Biol., 395, 2010
|
|
3FNV
 
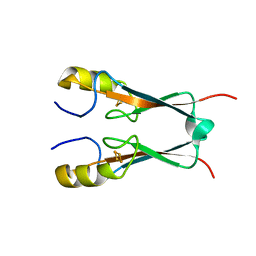 | | Crystal Structure of Miner1: The Redox-active 2Fe-2S Protein Causative in Wolfram Syndrome 2 | | Descriptor: | CDGSH iron sulfur domain-containing protein 2, FE2/S2 (INORGANIC) CLUSTER | | Authors: | Conlan, A.R, Axelrod, H.L, Cohen, A.E, Abresch, E.C, Yee, D, Zuris, J, Nechushtai, R, Jennings, P.A, Paddock, M.L. | | Deposit date: | 2008-12-26 | | Release date: | 2009-08-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of Miner1: The redox-active 2Fe-2S protein causative in Wolfram Syndrome 2.
J.Mol.Biol., 392, 2009
|
|
4LF1
 
 | | Hexameric Form II RuBisCO from Rhodopseudomonas palustris, activated and complexed with 2-CABP | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, Ribulose bisphosphate carboxylase | | Authors: | Chan, S, Satagopan, S, Sawaya, M.R, Eisenberg, D, Tabita, F.R, Perry, L.J. | | Deposit date: | 2013-06-26 | | Release date: | 2014-06-25 | | Last modified: | 2016-07-20 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structure-function studies with the unique hexameric form II ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) from Rhodopseudomonas palustris.
J.Biol.Chem., 289, 2014
|
|
3NSW
 
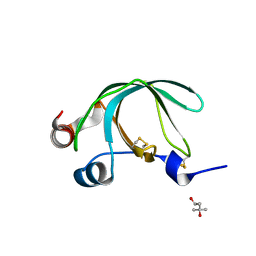 | | Crystal Structure of Ancylostoma ceylanicum Excretory-Secretory Protein 2 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Excretory-secretory protein 2 | | Authors: | Kucera, K, Modis, Y. | | Deposit date: | 2010-07-02 | | Release date: | 2011-05-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Ancylostoma ceylanicum Excretory-Secretory Protein 2 Adopts a Netrin-Like Fold and Defines a Novel Family of Nematode Proteins.
J.Mol.Biol., 408, 2011
|
|
4M9I
 
 | |
