1W7O
 
 | | cytochrome c3 from Desulfomicrobium baculatus | | Descriptor: | CYTOCHROME C3, HEME C | | Authors: | Coelho, A.V, Frazao, C, Matias, P.M, Carrondo, M.A. | | Deposit date: | 2004-09-07 | | Release date: | 2004-09-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Proton-Assisted Two-Electron Transfer in Natural Variants of Tetraheme Cytochromes from Desulfomicrobium Sp
J.Biol.Chem., 279, 2004
|
|
6XXI
 
 | | Crystal Structure of Human Deoxyhypusine Synthase in complex with NAD | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Wator, E, Wilk, P, Grudnik, P. | | Deposit date: | 2020-01-27 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.679 Å) | | Cite: | Half Way to Hypusine-Structural Basis for Substrate Recognition by Human Deoxyhypusine Synthase.
Biomolecules, 10, 2020
|
|
5A0M
 
 | | THE CRYSTAL STRUCTURE OF I-SCEI IN COMPLEX WITH ITS TARGET DNA IN THE PRESENCE OF MN | | Descriptor: | 5'-D(*CP*AP*CP*GP*CP*TP*AP*GP*GP*GP*AP*TP*AP*AP)-3', 5'-D(*CP*AP*GP*GP*GP*TP*AP*AP*TP*AP*CP)-3', 5'-D(*CP*CP*CP*TP*AP*GP*CP*GP*TP)-3', ... | | Authors: | Prieto, J, Redondo, P, Merino, N, Villate, M, Blanco, F.J, Molina, R. | | Deposit date: | 2015-04-21 | | Release date: | 2016-05-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the I-Scei Nuclease Complexed with its DsDNA Target and Three Catalytic Metal Ions.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
4V7Z
 
 | | Structure of the Thermus thermophilus 70S ribosome complexed with telithromycin. | | Descriptor: | 16S rRNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Bulkley, D.P, Innis, C.A, Blaha, G, Steitz, T.A. | | Deposit date: | 2010-08-18 | | Release date: | 2014-07-09 | | Last modified: | 2021-01-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Revisiting the structures of several antibiotics bound to the bacterial ribosome.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
1W8B
 
 | | Factor7 - 413 complex | | Descriptor: | (S)-[(R)-2-(4-BENZYLOXY-3-METHOXY-PHENYL)-2-(4-CARBAMIMIDOYL-PHENYLAMINO)-ACETYLAMINO]-PHENYL-ACETIC ACID, BLOOD COAGULATION FACTOR VIIA, CALCIUM ION | | Authors: | Ackermann, J, Alig, L, Banner, D.W, Boehm, H.-J, Groebke-Zbinden, K, Hilpert, K, Lave, T, Kuehne, H, Obst-Sander, U, Riederer, M.A, Stahl, M, Tschopp, T.B, Weber, L, Wessel, H.P. | | Deposit date: | 2004-09-17 | | Release date: | 2005-10-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Selective and Orally Bioavailable Phenylglycine Tissue Factor/Factor Viia Inhibitors
Bioorg.Med.Chem.Lett., 15, 2005
|
|
6R95
 
 | |
4WAD
 
 | | Crystal Structure of TarM with UDP-GlcNAc | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Glycosyl transferase, ... | | Authors: | Koc, C, Stehle, T, Xia, G, Peschel, A. | | Deposit date: | 2014-08-29 | | Release date: | 2015-02-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and Enzymatic Analysis of TarM Glycosyltransferase from Staphylococcus aureus Reveals an Oligomeric Protein Specific for the Glycosylation of Wall Teichoic Acid.
J.Biol.Chem., 290, 2015
|
|
6QXB
 
 | | NMR structure of peptide 7, characterized by a cis-4-amino-Pro residue, with a significant lower MIC on E. coli | | Descriptor: | PHE-VAL-CAP-TRP-PHE-SER-LYS-PHE-LEU-GLY-ARG-ILE-LEU-NH2 | | Authors: | Brancaccio, D, Carotenuto, A, Merlino, F, Grieco, P, Novellino, E. | | Deposit date: | 2019-03-07 | | Release date: | 2019-05-29 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The Outcomes of Decorated Prolines in the Discovery of Antimicrobial Peptides from Temporin-L.
Chemmedchem, 14, 2019
|
|
6R96
 
 | |
1N2R
 
 | | A natural selected dimorphism in HLA B*44 alters self, peptide reportoire and T cell recognition. | | Descriptor: | ACETIC ACID, Beta-2-microglobulin, HLA DPA*0201 PEPTIDE, ... | | Authors: | Macdonald, W.A, Purcell, A.W, Williams, D.S, Mifsud, N, Ely, L.K, Gorman, J.J, Clements, C.S, Kjer-Nielsen, L, Koelle, D.M, Brooks, A.G, Lovrecz, G.O, Lu, L, Rossjohn, J, McCluskey, J. | | Deposit date: | 2002-10-24 | | Release date: | 2004-03-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A naturally selected dimorphism within the HLA-B44 supertype alters class I structure, peptide repertoire, and T cell recognition.
J.Exp.Med., 198, 2003
|
|
6QXC
 
 | | NMR structure of peptide 8, characterized by a trans-4-cyclohexyl-Pro, with a dramatic reduction in activity on E. coli ATCC and lost effect on P. aeruginosa. | | Descriptor: | PHE-VAL-TCP-TRP-PHE-SER-LYS-PHE-LEU-GLY-ARG-ILE-LEU-NH2 | | Authors: | Brancaccio, D, Carotenuto, A, Merlino, F, Grieco, P, Novellino, E. | | Deposit date: | 2019-03-07 | | Release date: | 2019-05-29 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The Outcomes of Decorated Prolines in the Discovery of Antimicrobial Peptides from Temporin-L.
Chemmedchem, 14, 2019
|
|
6R5G
 
 | |
4WCI
 
 | | Crystal structure of the 1st SH3 domain from human CD2AP (CMS) in complex with a proline-rich peptide (aa 378-393) from human RIN3 | | Descriptor: | CD2-associated protein, Ras and Rab interactor 3, SULFATE ION | | Authors: | Rouka, E, Simister, P.C, Janning, M, Kirsch, K.H, Krojer, T, Knapp, S, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Feller, S.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-09-04 | | Release date: | 2015-08-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Differential Recognition Preferences of the Three Src Homology 3 (SH3) Domains from the Adaptor CD2-associated Protein (CD2AP) and Direct Association with Ras and Rab Interactor 3 (RIN3).
J.Biol.Chem., 290, 2015
|
|
4WDA
 
 | | Catalytic domain of mouse 2',3'-cyclic nucleotide 3'- phosphodiesterase, with mutation P296G, complexed with 2'-AMP | | Descriptor: | 2',3'-cyclic-nucleotide 3'-phosphodiesterase, ADENOSINE-2'-MONOPHOSPHATE | | Authors: | Myllykoski, M, Raasakka, A, Kursula, P. | | Deposit date: | 2014-09-08 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Determinants of ligand binding and catalytic activity in the myelin enzyme 2',3'-cyclic nucleotide 3'-phosphodiesterase.
Sci Rep, 5, 2015
|
|
3VFO
 
 | | crystal structure of HLA B*3508 LPEP157A, HLA mutant Ala157 | | Descriptor: | Beta-2-microglobulin, LPEP peptide from EBV, LPEPLPQGQLTAY, ... | | Authors: | Liu, Y.C, Rossjohn, J, Gras, S. | | Deposit date: | 2012-01-10 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Energetic Basis Underpinning T-cell Receptor Recognition of a Super-bulged Peptide Bound to a Major Histocompatibility Complex Class I Molecule.
J.Biol.Chem., 287, 2012
|
|
6RGI
 
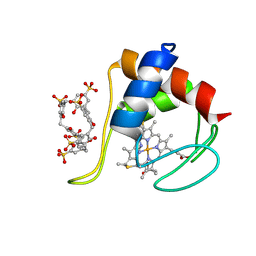 | | Partially unfolded cytochrome c in complex with sulfonatocalix[6]arene | | Descriptor: | Cytochrome c iso-1, HEME C, IMIDAZOLE, ... | | Authors: | Engilberge, S, Rennie, M.L, Crowley, P.B. | | Deposit date: | 2019-04-16 | | Release date: | 2019-07-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Calixarene capture of partially unfolded cytochrome c.
Febs Lett., 593, 2019
|
|
6XF2
 
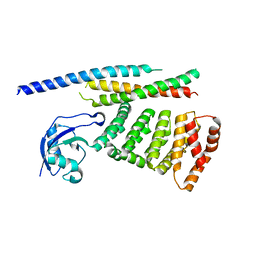 | |
6RGU
 
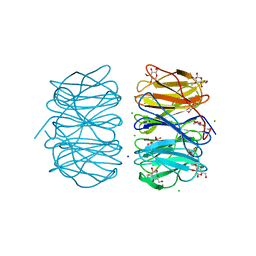 | | Photorhabdus asymbiotica lectin PHL in complex with 3-O-methyl-D-glucose | | Descriptor: | 3-O-methyl-beta-D-glucopyranose, CHLORIDE ION, Lectin PHL, ... | | Authors: | Houser, J, Fujdiarova, E, Jancarikova, G, Wimmerova, M. | | Deposit date: | 2019-04-17 | | Release date: | 2020-07-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Heptabladed beta-propeller lectins PLL2 and PHL from Photorhabdus spp. recognize O-methylated sugars and influence the host immune system.
Febs J., 288, 2021
|
|
4WC9
 
 | | Catalytic domain of mouse 2',3'-cyclic nucleotide 3'- phosphodiesterase, with mutation F235L | | Descriptor: | 2',3'-cyclic-nucleotide 3'-phosphodiesterase | | Authors: | Myllykoski, M, Raasakka, A, Kursula, P. | | Deposit date: | 2014-09-04 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Determinants of ligand binding and catalytic activity in the myelin enzyme 2',3'-cyclic nucleotide 3'-phosphodiesterase.
Sci Rep, 5, 2015
|
|
6XKW
 
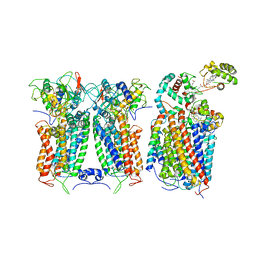 | | R. capsulatus CIII2CIV bipartite super-complex (SC-2A) with CcoH/cy | | Descriptor: | COPPER (II) ION, Cbb3-type cytochrome c oxidase subunit CcoP, Cytochrome b, ... | | Authors: | Steimle, S, Van Eeuwen, T, Ozturk, Y, Kim, H.J, Braitbard, M, Selamoglu, N, Garcia, B.A, Schneidman-Duhovny, D, Murakami, K, Daldal, F. | | Deposit date: | 2020-06-27 | | Release date: | 2020-12-30 | | Last modified: | 2021-03-03 | | Method: | ELECTRON MICROSCOPY (5.2 Å) | | Cite: | Cryo-EM structures of engineered active bc 1 -cbb 3 type CIII 2 CIV super-complexes and electronic communication between the complexes.
Nat Commun, 12, 2021
|
|
1N2F
 
 | | CRYSTAL STRUCTURE OF P. AERUGINOSA OHR | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Organic Hydroperoxide Resistance Protein | | Authors: | Lesniak, J, Barton, W.A, Nikolov, D.B. | | Deposit date: | 2002-10-22 | | Release date: | 2002-12-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural and functional characterization of the Pseudomonas hydroperoxide resistance protein Ohr
Embo J., 21, 2002
|
|
1MSE
 
 | | SOLUTION STRUCTURE OF A SPECIFIC DNA COMPLEX OF THE MYB DNA-BINDING DOMAIN WITH COOPERATIVE RECOGNITION HELICES | | Descriptor: | C-Myb DNA-Binding Domain, DNA (5'-D(*AP*TP*GP*TP*GP*TP*GP*TP*CP*AP*GP*TP*TP*AP*GP*G)-3'), DNA (5'-D(*CP*CP*TP*AP*AP*CP*TP*GP*AP*CP*AP*CP*AP*CP*AP*T)-3') | | Authors: | Ogata, K, Morikawa, S, Nakamura, H, Sekikawa, A, Inoue, T, Kanai, H, Sarai, A, Ishii, S, Nishimura, Y. | | Deposit date: | 1995-01-24 | | Release date: | 1995-03-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a specific DNA complex of the Myb DNA-binding domain with cooperative recognition helices.
Cell(Cambridge,Mass.), 79, 1994
|
|
6XI7
 
 | | Crystal Structure of wild-type KRAS (GMPPNP-bound) in complex with RAS-binding domain (RBD) and cysteine-rich domain (CRD) of RAF1/CRAF (crystal form I) | | Descriptor: | CHLORIDE ION, Isoform 2B of GTPase KRas, MAGNESIUM ION, ... | | Authors: | Chan, A.H, Tran, T.H, Dharmaiah, S, Simanshu, D.K. | | Deposit date: | 2020-06-19 | | Release date: | 2021-01-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | KRAS interaction with RAF1 RAS-binding domain and cysteine-rich domain provides insights into RAS-mediated RAF activation.
Nat Commun, 12, 2021
|
|
1VFR
 
 | | THE MAJOR NAD(P)H:FMN OXIDOREDUCTASE FROM VIBRIO FISCHERI | | Descriptor: | FLAVIN MONONUCLEOTIDE, NAD(P)H:FMN OXIDOREDUCTASE | | Authors: | Koike, H, Sasaki, H, Kobori, T, Zenno, S, Saigo, K, Murphy, M.E.P, Adman, E.T, Tanokura, M. | | Deposit date: | 1998-01-09 | | Release date: | 1999-02-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1.8 A crystal structure of the major NAD(P)H:FMN oxidoreductase of a bioluminescent bacterium, Vibrio fischeri: overall structure, cofactor and substrate-analog binding, and comparison with related flavoproteins.
J.Mol.Biol., 280, 1998
|
|
6RI7
 
 | | Cryo-EM structure of E. coli RNA polymerase elongation complex bound to GreB transcription factor | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Abdelkareem, M, Saint-Andre, C, Takacs, M, Papai, G, Crucifix, C, Guo, X, Ortiz, J, Weixlbaumer, A. | | Deposit date: | 2019-04-23 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural Basis of Transcription: RNA Polymerase Backtracking and Its Reactivation.
Mol.Cell, 75, 2019
|
|
