8AGC
 
 | | Structure of yeast oligosaccharylransferase complex with lipid-linked oligosaccharide and non-acceptor peptide bound | | Descriptor: | 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-[3,6-bis(dimethylamino)xanthen-9-yl]-5-methanoyl-benzoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ramirez, A.S, de Capitani, M, Pesciullesi, G, Kowal, J, Bloch, J.S, Irobalieva, R.N, Aebi, M, Reymond, J.L, Locher, K.P. | | Deposit date: | 2022-07-19 | | Release date: | 2022-12-07 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular basis for glycan recognition and reaction priming of eukaryotic oligosaccharyltransferase.
Nat Commun, 13, 2022
|
|
5WNJ
 
 | |
2G73
 
 | | Y104F mutant type 1 IPP isomerase complex with EIPP | | Descriptor: | 4-HYDROXY-3-METHYL BUTYL DIPHOSPHATE, Isopentenyl-diphosphate delta-isomerase, MAGNESIUM ION, ... | | Authors: | de Ruyck, J, Wouters, J. | | Deposit date: | 2006-02-27 | | Release date: | 2006-04-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural role for Tyr-104 in Escherichia coli isopentenyl-diphosphate isomerase: site-directed mutagenesis, enzymology, and protein crystallography.
J.Biol.Chem., 281, 2006
|
|
7ZLJ
 
 | | Cryo-EM structure of C-mannosyltransferase CeDPY19, in ternary complex with Dol25-P-C-Man and acceptor peptide, bound to CMT2-Fab and anti-Fab nanobody | | Descriptor: | Anti-Fab nanobody, C-mannosyltransferase dpy-19, CMT2-Fab heavy chain, ... | | Authors: | Bloch, J.S, Mao, R, Mukherjee, S, Boilevin, J, Irobalieva, R, Darbre, T, Reymond, J.L, Kossiakoff, A.A, Goddard-Borger, E.D, Locher, K.P. | | Deposit date: | 2022-04-15 | | Release date: | 2023-01-11 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.63 Å) | | Cite: | Structure, sequon recognition and mechanism of tryptophan C-mannosyltransferase.
Nat.Chem.Biol., 19, 2023
|
|
7ZLI
 
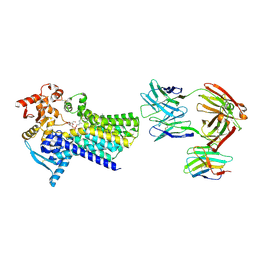 | | Cryo-EM structure of C-mannosyltransferase CeDPY19, in complex with Dol25-P-Man and bound to CMT2-Fab and anti-Fab nanobody | | Descriptor: | Anti-Fab nanobody, C-mannosyltransferase dpy-19, CMT2-Fab heavy chain, ... | | Authors: | Bloch, J.S, Mukherjee, S, Boilevin, J, Irobalieva, R, Darbre, T, Reymond, J.L, Kossiakoff, A.A, Goddard-Borger, E.D, Locher, K.P. | | Deposit date: | 2022-04-15 | | Release date: | 2023-01-11 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Structure, sequon recognition and mechanism of tryptophan C-mannosyltransferase.
Nat.Chem.Biol., 19, 2023
|
|
5WNK
 
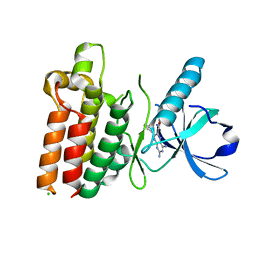 | | Crystal structure of murine receptor-interacting protein 4 (Ripk4) D143N bound to TG100-115 | | Descriptor: | 3,3'-(2,4-diaminopteridine-6,7-diyl)diphenol, CHLORIDE ION, Receptor-interacting serine/threonine-protein kinase 4 | | Authors: | Huang, C.S, Hymowitz, S.G. | | Deposit date: | 2017-08-01 | | Release date: | 2018-05-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Crystal Structure of Ripk4 Reveals Dimerization-Dependent Kinase Activity.
Structure, 26, 2018
|
|
2TGD
 
 | |
3TMO
 
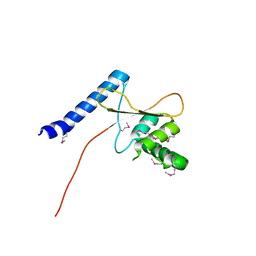 | | The catalytic domain of human deubiquitinase DUBA | | Descriptor: | OTU domain-containing protein 5 | | Authors: | Yin, J, Bosanac, I, Ma, X, Hymowitz, S, Starovasnik, M, Cochran, A. | | Deposit date: | 2011-08-31 | | Release date: | 2012-01-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Phosphorylation-dependent activity of the deubiquitinase DUBA.
Nat.Struct.Mol.Biol., 19, 2012
|
|
1VAR
 
 | |
4MNE
 
 | | Crystal structure of the BRAF:MEK1 complex | | Descriptor: | 7-fluoro-3-[(2-fluoro-4-iodophenyl)amino]-N-{[(2S)-2-hydroxypropyl]oxy}furo[3,2-c]pyridine-2-carboxamide, CHLORIDE ION, Dual specificity mitogen-activated protein kinase kinase 1, ... | | Authors: | Sudhamsu, J, Haling, J.R, Morales, T, Brandhuber, B, Hymowitz, S.G. | | Deposit date: | 2013-09-10 | | Release date: | 2014-06-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8483 Å) | | Cite: | Structure of the BRAF-MEK Complex Reveals a Kinase Activity Independent Role for BRAF in MAPK Signaling.
Cancer Cell, 26, 2014
|
|
4MNF
 
 | | Crystal structure of BRAF-V600E bound to GDC0879 | | Descriptor: | 2-{4-[(1E)-1-(hydroxyimino)-2,3-dihydro-1H-inden-5-yl]-3-(pyridin-4-yl)-1H-pyrazol-1-yl}ethanol, CHLORIDE ION, Serine/threonine-protein kinase B-raf | | Authors: | Sudhamsu, J, Haling, J.R, Morales, T, Brandhuber, B, Hymowitz, S.G. | | Deposit date: | 2013-09-10 | | Release date: | 2014-06-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.802 Å) | | Cite: | Structure of the BRAF-MEK Complex Reveals a Kinase Activity Independent Role for BRAF in MAPK Signaling.
Cancer Cell, 26, 2014
|
|
3SQE
 
 | |
3T6P
 
 | | IAP antagonist-induced conformational change in cIAP1 promotes E3 ligase activation via dimerization | | Descriptor: | Baculoviral IAP repeat-containing protein 2, ZINC ION | | Authors: | Dueber, E.C, Schoeffler, A.J, Lingel, A, Elliott, M, Fedorova, A.V, Giannetti, A.M, Zobel, K, Maurer, B, Varfolomeev, E, Wu, P, Wallweber, H, Hymowitz, S, Deshayes, K, Vucic, D, Fairbrother, W.J. | | Deposit date: | 2011-07-28 | | Release date: | 2011-11-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.897 Å) | | Cite: | Antagonists induce a conformational change in cIAP1 that promotes autoubiquitination.
Science, 334, 2011
|
|
4MXW
 
 | | Structure of heterotrimeric lymphotoxin LTa1b2 bound to lymphotoxin beta receptor LTbR and anti-LTa Fab | | Descriptor: | Lymphotoxin-alpha, Lymphotoxin-beta, Tumor necrosis factor receptor superfamily member 3, ... | | Authors: | Sudhamsu, J, Yin, J.P, Hymowitz, S.G. | | Deposit date: | 2013-09-26 | | Release date: | 2013-11-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Dimerization of LT beta R by LT alpha 1 beta 2 is necessary and sufficient for signal transduction.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4NRV
 
 | | Crystal Structure of non-edited human NEIL1 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Endonuclease 8-like 1 | | Authors: | Prakash, A, Doublie, S. | | Deposit date: | 2013-11-27 | | Release date: | 2014-01-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Genome and cancer single nucleotide polymorphisms of the human NEIL1 DNA glycosylase: Activity, structure, and the effect of editing.
Dna Repair, 14, 2014
|
|
8QXB
 
 | | TDP-43 amyloid fibrils: Morphology-2 | | Descriptor: | TAR DNA-binding protein 43 | | Authors: | Sharma, K, Shenoy, J, Loquet, A, Schmidt, M, Faendrich, M. | | Deposit date: | 2023-10-24 | | Release date: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.86 Å) | | Cite: | Cryo-EM observation of the amyloid key structure of polymorphic TDP-43 amyloid fibrils.
Nat Commun, 15, 2024
|
|
8QXA
 
 | | TDP-43 amyloid fibrils: Morphology-1b | | Descriptor: | TAR DNA-binding protein 43 | | Authors: | Sharma, K, Shenoy, J, Loquet, A, Schmidt, M, Faendrich, M. | | Deposit date: | 2023-10-24 | | Release date: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (4.05 Å) | | Cite: | Cryo-EM observation of the amyloid key structure of polymorphic TDP-43 amyloid fibrils.
Nat Commun, 15, 2024
|
|
8QX9
 
 | | TDP-43 amyloid fibrils: Morphology-1a | | Descriptor: | TAR DNA-binding protein 43 | | Authors: | Sharma, K, Shenoy, J, Loquet, A, Schmidt, M, Faendrich, M. | | Deposit date: | 2023-10-24 | | Release date: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.76 Å) | | Cite: | Cryo-EM observation of the amyloid key structure of polymorphic TDP-43 amyloid fibrils.
Nat Commun, 15, 2024
|
|
3SQH
 
 | |
4NRW
 
 | | MvNei1-G86D | | Descriptor: | 5'-D(*CP*GP*TP*CP*CP*AP*(3DR)P*GP*TP*CP*TP*AP*C)-3', 5'-D(*GP*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*G)-3', formamidopyrimidine-DNA glycosylase | | Authors: | Prakash, A, Doublie, S. | | Deposit date: | 2013-11-27 | | Release date: | 2014-01-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.845 Å) | | Cite: | Genome and cancer single nucleotide polymorphisms of the human NEIL1 DNA glycosylase: Activity, structure, and the effect of editing.
Dna Repair, 14, 2014
|
|
8OXI
 
 | |
8OXH
 
 | |
8OXK
 
 | |
8OXL
 
 | |
8OXJ
 
 | |
