8CUP
 
 | | X-ray crystal structure of ADC-33 in complex with sulfonamidoboronic acid 6d | | Descriptor: | 3-[(4S)-4-ethyl-5,7,7-trihydroxy-2,2,7-trioxo-6-oxa-2lambda~6~-thia-3-aza-7lambda~5~-phospha-5-boraheptan-1-yl]benzoic acid, Beta-lactamase | | Authors: | Fernando, M.C, Wallar, B.J, Powers, R.A. | | Deposit date: | 2022-05-17 | | Release date: | 2023-04-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Sulfonamidoboronic Acids as "Cross-Class" Inhibitors of an Expanded-Spectrum Class C Cephalosporinase, ADC-33, and a Class D Carbapenemase, OXA-24/40: Strategic Compound Design to Combat Resistance in Acinetobacter baumannii .
Antibiotics, 12, 2023
|
|
8CUQ
 
 | | X-ray crystal structure of ADC-33 in complex with sulfonamidoboronic acid 6e | | Descriptor: | 3-[(4R)-4-ethyl-5,7,7-trihydroxy-2,2,7-trioxo-6-oxa-2lambda~6~-thia-3-aza-7lambda~5~-phospha-5-boraheptan-1-yl]benzoic acid, Beta-lactamase | | Authors: | Fernando, M.C, Wallar, B.J, Powers, R.A. | | Deposit date: | 2022-05-17 | | Release date: | 2023-04-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Sulfonamidoboronic Acids as "Cross-Class" Inhibitors of an Expanded-Spectrum Class C Cephalosporinase, ADC-33, and a Class D Carbapenemase, OXA-24/40: Strategic Compound Design to Combat Resistance in Acinetobacter baumannii .
Antibiotics, 12, 2023
|
|
8DC1
 
 | |
8DDZ
 
 | | TEM-1 beta-lactamase A237Y | | Descriptor: | Beta-lactamase TEM | | Authors: | Ji, Z, Boxer, S.G, Mathews, I.I. | | Deposit date: | 2022-06-19 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Protein Electric Fields Enable Faster and Longer-Lasting Covalent Inhibition of beta-Lactamases.
J.Am.Chem.Soc., 144, 2022
|
|
8DE0
 
 | | TEM-1 beta-lactamase covalently bound to avibactam | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, Beta-lactamase TEM | | Authors: | Ji, Z, Boxer, S.G, Mathews, I.I. | | Deposit date: | 2022-06-19 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Protein Electric Fields Enable Faster and Longer-Lasting Covalent Inhibition of beta-Lactamases.
J.Am.Chem.Soc., 144, 2022
|
|
8DE1
 
 | |
8DE2
 
 | | TEM-1 beta-lactamase A237Y mutant covalently bound to avibactam, a room temperature structure | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, Beta-lactamase TEM | | Authors: | Ji, Z, Boxer, S.G, Mathews, I.I. | | Deposit date: | 2022-06-19 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Protein Electric Fields Enable Faster and Longer-Lasting Covalent Inhibition of beta-Lactamases.
J.Am.Chem.Soc., 144, 2022
|
|
8DI7
 
 | | CMY-2 | | Descriptor: | Beta-lactamase, SULFATE ION | | Authors: | Ahmadvand, P, Call, D.R. | | Deposit date: | 2022-06-28 | | Release date: | 2023-06-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural characterization of CMY-2 and its interactions with ampicillin and the cephalosporins - ceftiofur, DFC, DFC-dimer, DFC-cysteine, and nitrocefin
To Be Published
|
|
8DOD
 
 | | Beta-lactamase CTX-M-14 S130A | | Descriptor: | Beta-lactamase, POTASSIUM ION | | Authors: | Lu, S, Palzkill, T. | | Deposit date: | 2022-07-12 | | Release date: | 2023-04-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Mutagenesis and structural analysis reveal the CTX-M beta-lactamase active site is optimized for cephalosporin catalysis and drug resistance.
J.Biol.Chem., 299, 2023
|
|
8DOE
 
 | | Crystal Structure of CTX-M-14 N106A | | Descriptor: | Beta-lactamase | | Authors: | Lu, S, Palzkill, T. | | Deposit date: | 2022-07-12 | | Release date: | 2023-04-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mutagenesis and structural analysis reveal the CTX-M beta-lactamase active site is optimized for cephalosporin catalysis and drug resistance.
J.Biol.Chem., 299, 2023
|
|
8DON
 
 | | Beta-lactamase CTX-M-14 T215A | | Descriptor: | Beta-lactamase, PHOSPHATE ION | | Authors: | Lu, S, Palzkill, T. | | Deposit date: | 2022-07-13 | | Release date: | 2023-04-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Mutagenesis and structural analysis reveal the CTX-M beta-lactamase active site is optimized for cephalosporin catalysis and drug resistance.
J.Biol.Chem., 299, 2023
|
|
8DP4
 
 | | Beta-lactamase CTX-M-14 T235A | | Descriptor: | Beta-lactamase | | Authors: | Lu, S, Palzkill, T. | | Deposit date: | 2022-07-14 | | Release date: | 2023-04-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.402 Å) | | Cite: | Mutagenesis and structural analysis reveal the CTX-M beta-lactamase active site is optimized for cephalosporin catalysis and drug resistance.
J.Biol.Chem., 299, 2023
|
|
8DPQ
 
 | | Beta-lactamase CTX-M-14 N170A | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase | | Authors: | Lu, S, Neetu, N, Palzkill, T. | | Deposit date: | 2022-07-15 | | Release date: | 2023-04-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Mutagenesis and structural analysis reveal the CTX-M beta-lactamase active site is optimized for cephalosporin catalysis and drug resistance.
J.Biol.Chem., 299, 2023
|
|
8EBI
 
 | |
8EBR
 
 | |
8EC4
 
 | |
8ECF
 
 | |
8EHH
 
 | | Crystal structure of the class A extended-spectrum beta-lactamase CTX-M-96 in complex with relebactam at 1.03 Angstrom resolution | | Descriptor: | (2S,5R)-1-formyl-N-(piperidin-4-yl)-5-[(sulfooxy)amino]piperidine-2-carboxamide, Beta-lactamase | | Authors: | Power, P, Ghiglione, B, Bonomo, R.A, Rodriguez, M.M, Gutkind, G, Klinke, S. | | Deposit date: | 2022-09-14 | | Release date: | 2023-09-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Biochemical and structural evidences of the activity of relebactam as inhibitor of the extended-spectrum beta-lactamase CTX-M-96.
To be published
|
|
8EHU
 
 | | Crystal structure of the environmental CRH-1 class A carbapenemase at 1.1 Angstrom resolution | | Descriptor: | Beta-lactamase | | Authors: | Power, P, Brunetti, F, Ghiglione, B, Guardabassi, L, Gutkind, G, Klinke, S. | | Deposit date: | 2022-09-14 | | Release date: | 2023-05-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Biochemical and Structural Characterization of CRH-1, a Carbapenemase from Chromobacterium haemolyticum Related to KPC beta-Lactamases.
Antimicrob.Agents Chemother., 67, 2023
|
|
8EK9
 
 | | Crystal structure of the class A carbapenemase CRH-1 in complex with avibactam at 1.4 Angstrom resolution | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, Beta-lactamase | | Authors: | Power, P, Brunetti, F, Ghiglione, B, Guardabassi, L, Gutkind, G, Klinke, S. | | Deposit date: | 2022-09-20 | | Release date: | 2023-05-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Biochemical and Structural Characterization of CRH-1, a Carbapenemase from Chromobacterium haemolyticum Related to KPC beta-Lactamases.
Antimicrob.Agents Chemother., 67, 2023
|
|
8ELA
 
 | | CTX-M-14 beta-lactamase mutant - N132A w MES | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-lactamase, CHLORIDE ION, ... | | Authors: | Lu, S, Palzkill, T, Hu, L, Prasad, B.V.V. | | Deposit date: | 2022-09-23 | | Release date: | 2023-04-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mutagenesis and structural analysis reveal the CTX-M beta-lactamase active site is optimized for cephalosporin catalysis and drug resistance.
J.Biol.Chem., 299, 2023
|
|
8ELB
 
 | | CTX-M-14 beta-lactamase mutant- N132A | | Descriptor: | Beta-lactamase, DI(HYDROXYETHYL)ETHER | | Authors: | Lu, S, Neetu, N, Palzkill, T. | | Deposit date: | 2022-09-23 | | Release date: | 2023-04-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mutagenesis and structural analysis reveal the CTX-M beta-lactamase active site is optimized for cephalosporin catalysis and drug resistance.
J.Biol.Chem., 299, 2023
|
|
8EO5
 
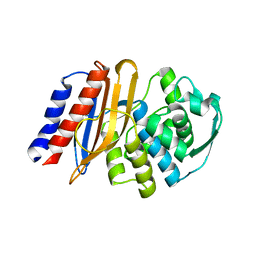 | | Crystal structure of the class A beta-lactamase precursor LRA-5 from an Alaskan soil metagenome at 1.8 Angstrom resolution | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, LRA-5 | | Authors: | Power, P, D'Amico Gonzalez, G, Centron, D, Gutkind, G, Handelsman, J, Klinke, S. | | Deposit date: | 2022-10-02 | | Release date: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Playing beta-Lactamase Evolution: Metagenomic Class A beta-Lactamase LRA-5 is an Inactive Enzyme Capable of Rendering an Active beta-Lactamase by Introduction of Y69Q and V166E Substitutions
to be published
|
|
8EO6
 
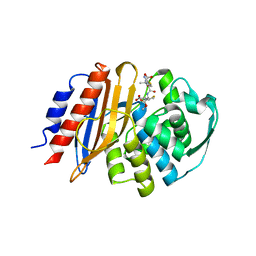 | | Crystal structure of metagenomic class A beta-lactamase precursor LRA-5 in complex with ceftazidime at 2.35 Angstrom resolution | | Descriptor: | ACYLATED CEFTAZIDIME, LRA-5 | | Authors: | Power, P, D'Amico Gonzalez, G, Centron, D, Gutkind, G, Handelsman, J, Klinke, S. | | Deposit date: | 2022-10-02 | | Release date: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Playing beta-Lactamase Evolution: Metagenomic Class A beta-Lactamase LRA-5 is an Inactive Enzyme Capable of Rendering an Active beta-Lactamase by Introduction of Y69Q and V166E Substitutions
to be published
|
|
8EO7
 
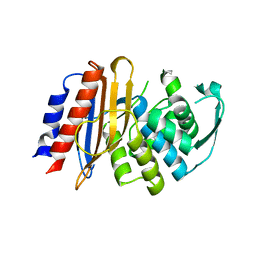 | | Crystal structure of metagenomic beta-lactamase LRA-5 Y69Q/V166E mutant at 2.15 Angstrom resolution | | Descriptor: | beta-lactamase | | Authors: | Power, P, D'Amico Gonzalez, G, Centron, D, Gutkind, G, Handelsman, J, Klinke, S. | | Deposit date: | 2022-10-02 | | Release date: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Playing beta-Lactamase Evolution: Metagenomic Class A beta-Lactamase LRA-5 is an Inactive Enzyme Capable of Rendering an Active beta-Lactamase by Introduction of Y69Q and V166E Substitutions
to be published
|
|
