4V40
 
 | | BETA-GALACTOSIDASE | | Descriptor: | BETA-GALACTOSIDASE, MAGNESIUM ION | | Authors: | Jacobson, R.H, Zhang, X, Dubose, R.F, Matthews, B.W. | | Deposit date: | 1994-07-18 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Three-dimensional structure of beta-galactosidase from E. coli.
Nature, 369, 1994
|
|
6O2O
 
 | |
3E3F
 
 | |
6NY5
 
 | |
5K7H
 
 | | Crystal structure of AibR in complex with the effector molecule isovaleryl coenzyme A | | Descriptor: | CHLORIDE ION, Isovaleryl-coenzyme A, NICKEL (II) ION, ... | | Authors: | Bock, T, Volz, C, Mueller, R, Blankenfeldt, W. | | Deposit date: | 2016-05-26 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The AibR-isovaleryl coenzyme A regulator and its DNA binding site - a model for the regulation of alternative de novo isovaleryl coenzyme A biosynthesis in Myxococcus xanthus.
Nucleic Acids Res., 45, 2017
|
|
8DO5
 
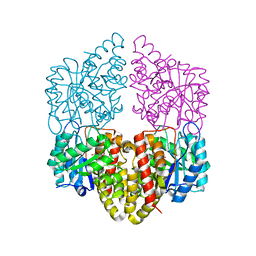 | | Crystal structure of NahE in complex with intermediate (R)-4-hydroxy-4-(2-hydroxyphenyl)-2-iminobutanoate | | Descriptor: | (4R)-4-hydroxy-4-(2-hydroxyphenyl)butanoic acid, DIMETHYL SULFOXIDE, Trans-ohydrobenzylidenepyruvate hydratase aldolase | | Authors: | LeVieux, J.A, Hardtke, H.A, Zhang, Y.J. | | Deposit date: | 2022-07-12 | | Release date: | 2022-12-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A mutagenic analysis of NahE, a hydratase-aldolase in the naphthalene degradative pathway.
Arch.Biochem.Biophys., 733, 2023
|
|
4WF7
 
 | | Crystal structures of trehalose synthase from Deinococcus radiodurans reveal that a closed conformation is involved in the intramolecular isomerization catalysis | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Wang, Y.L, Chow, S.Y, Lin, Y.T, Hsieh, Y.C, Lee, G.C, Liaw, S.H. | | Deposit date: | 2014-09-13 | | Release date: | 2014-12-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structures of trehalose synthase from Deinococcus radiodurans reveal that a closed conformation is involved in catalysis of the intramolecular isomerization.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
8E2R
 
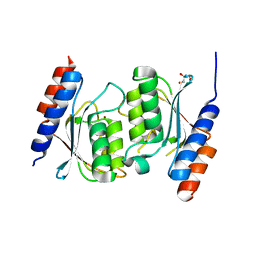 | | Crystal structure of TadAC-1.14 | | Descriptor: | GLYCEROL, ZINC ION, tRNA-specific adenosine deaminase 1.14 | | Authors: | Feliciano, P.R, Lee, S.J, Ciaramella, G. | | Deposit date: | 2022-08-15 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Improved cytosine base editors generated from TadA variants.
Nat.Biotechnol., 41, 2023
|
|
8E2P
 
 | |
8DQV
 
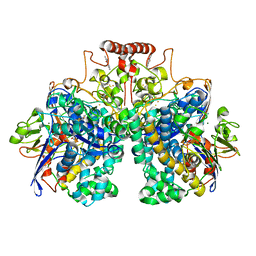 | | The 1.52 angstrom CryoEM structure of the [NiFe]-hydrogenase Huc from Mycobacterium smegmatis - catalytic dimer (Huc2S2L) | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, Hydrogenase-2, ... | | Authors: | Grinter, R, Venugopal, H, Kropp, A, Greening, C. | | Deposit date: | 2022-07-20 | | Release date: | 2023-01-04 | | Last modified: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (1.52 Å) | | Cite: | Structural basis for bacterial energy extraction from atmospheric hydrogen.
Nature, 615, 2023
|
|
8E2S
 
 | |
8E2Q
 
 | | Crystal structure of TadAC-1.17 in a complex with ssDNA | | Descriptor: | DNA (5'-D(P*GP*CP*GP*GP*CP*TP*(D8A)P*CP*GP*GP*A)-3'), GLYCEROL, ZINC ION, ... | | Authors: | Feliciano, P.R, Lee, S.J, Ciaramella, G. | | Deposit date: | 2022-08-15 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Improved cytosine base editors generated from TadA variants.
Nat.Biotechnol., 41, 2023
|
|
8E73
 
 | | Vigna radiata supercomplex I+III2 (full bridge) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, CARDIOLIPIN, ... | | Authors: | Maldonado, M, Letts, J.A. | | Deposit date: | 2022-08-23 | | Release date: | 2023-01-11 | | Last modified: | 2023-02-08 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Plant-specific features of respiratory supercomplex I + III 2 from Vigna radiata.
Nat.Plants, 9, 2023
|
|
6O65
 
 | |
8DWT
 
 | | SPOP W22R Form 2 | | Descriptor: | Speckle-type POZ protein | | Authors: | Cuneo, M.J, Mittag, T, O'Flynn, B, Lo, Y.H. | | Deposit date: | 2022-08-02 | | Release date: | 2023-01-18 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Higher-order SPOP assembly reveals a basis for cancer mutant dysregulation.
Mol.Cell, 83, 2023
|
|
5KGN
 
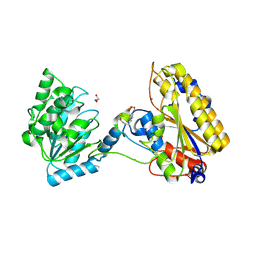 | | 1.95A resolution structure of independent phosphoglycerate mutase from C. elegans in complex with a macrocyclic peptide inhibitor (2d) | | Descriptor: | 2,3-bisphosphoglycerate-independent phosphoglycerate mutase, CHLORIDE ION, GLYCEROL, ... | | Authors: | Lovell, S, Mehzabeen, N, Battaile, K.P, Yu, H, Dranchak, P, MacArthur, R, Li, Z, Carlow, T, Suga, H, Inglese, J. | | Deposit date: | 2016-06-13 | | Release date: | 2017-04-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Macrocycle peptides delineate locked-open inhibition mechanism for microorganism phosphoglycerate mutases.
Nat Commun, 8, 2017
|
|
8DWV
 
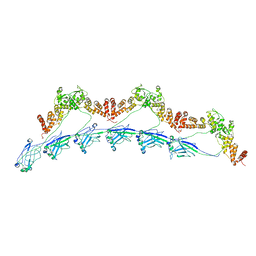 | | Full-length wild type SPOP | | Descriptor: | Speckle-type POZ protein | | Authors: | Cuneo, M.J, Mittag, T, O'Flynn, B, Lo, Y.H. | | Deposit date: | 2022-08-02 | | Release date: | 2023-01-18 | | Last modified: | 2023-03-22 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Higher-order SPOP assembly reveals a basis for cancer mutant dysregulation.
Mol.Cell, 83, 2023
|
|
8DWS
 
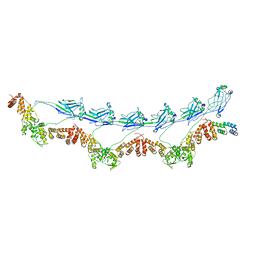 | | Full-length E47K SPOP | | Descriptor: | Speckle-type POZ protein | | Authors: | Cuneo, M.J, Mittag, T, O'Flynn, B, Lo, Y.H. | | Deposit date: | 2022-08-02 | | Release date: | 2023-01-18 | | Last modified: | 2023-03-22 | | Method: | ELECTRON MICROSCOPY (3.73 Å) | | Cite: | Higher-order SPOP assembly reveals a basis for cancer mutant dysregulation.
Mol.Cell, 83, 2023
|
|
8DWU
 
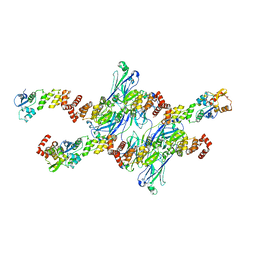 | | SPOP W22R Form 1 | | Descriptor: | Speckle-type POZ protein | | Authors: | Cuneo, M.J, Mittag, T, O'Flynn, B. | | Deposit date: | 2022-08-02 | | Release date: | 2023-01-18 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Higher-order SPOP assembly reveals a basis for cancer mutant dysregulation.
Mol.Cell, 83, 2023
|
|
6OKU
 
 | |
5JR7
 
 | | Crystal structure of an ACRDYS heterodimer [RIa(92-365):C] of PKA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase type I-alpha regulatory subunit | | Authors: | Bruystens, J.G.H, Wu, J, Taylor, S.S. | | Deposit date: | 2016-05-05 | | Release date: | 2017-03-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.56 Å) | | Cite: | Structure of a PKA RI alpha Recurrent Acrodysostosis Mutant Explains Defective cAMP-Dependent Activation.
J. Mol. Biol., 428, 2016
|
|
5JXZ
 
 | | A low magnesium structure of the isochorismate synthase, EntC | | Descriptor: | (3R,4R)-3-[(1-carboxyethenyl)oxy]-4-hydroxycyclohexa-1,5-diene-1-carboxylic acid, (5S,6S)-5-[(1-carboxyethenyl)oxy]-6-hydroxycyclohexa-1,3-diene-1-carboxylic acid, Isochorismate synthase EntC, ... | | Authors: | Meneely, K.M, Sundlov, J.A, Gulick, A.M, Lamb, A.L. | | Deposit date: | 2016-05-13 | | Release date: | 2016-07-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | An Open and Shut Case: The Interaction of Magnesium with MST Enzymes.
J.Am.Chem.Soc., 138, 2016
|
|
5JZD
 
 | | A re-refinement of the isochorismate synthase EntC | | Descriptor: | (5S,6S)-5-[(1-carboxyethenyl)oxy]-6-hydroxycyclohexa-1,3-diene-1-carboxylic acid, Isochorismate synthase EntC, MAGNESIUM ION | | Authors: | Meneely, K.M, Lamb, A.L. | | Deposit date: | 2016-05-16 | | Release date: | 2016-07-20 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | An Open and Shut Case: The Interaction of Magnesium with MST Enzymes.
J.Am.Chem.Soc., 138, 2016
|
|
1TD2
 
 | | Crystal Structure of the PdxY Protein from Escherichia coli | | Descriptor: | 3-HYDROXY-5-(HYDROXYMETHYL)-2-METHYLISONICOTINALDEHYDE, Pyridoxamine kinase, SULFATE ION | | Authors: | Safo, M.K, Musayev, F.N, Hunt, S, di Salvo, M, Scarsdale, N, Schirch, V. | | Deposit date: | 2004-05-21 | | Release date: | 2004-07-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Crystal structure of the PdxY Protein from Escherichia coli
J.Bacteriol., 186, 2004
|
|
6ON5
 
 | |
