3V79
 
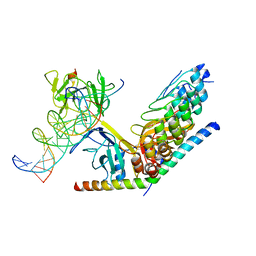 | | Structure of human Notch1 transcription complex including CSL, RAM, ANK, and MAML-1 on HES-1 promoter DNA sequence | | Descriptor: | DNA 5'-D(*GP*TP*TP*AP*CP*TP*GP*TP*GP*GP*GP*AP*AP*AP*GP*AP*AP*A)-3', DNA 5'-D(*TP*TP*TP*CP*TP*TP*TP*CP*CP*CP*AP*CP*AP*GP*TP*AP*AP*C)-3', Mastermind-like protein 1, ... | | Authors: | Nam, Y, Sliz, P, Blacklow, S. | | Deposit date: | 2011-12-20 | | Release date: | 2012-02-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.85 Å) | | Cite: | Conformational Locking upon Cooperative Assembly of Notch Transcription Complexes.
Structure, 20, 2012
|
|
7KPX
 
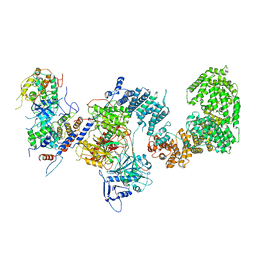 | | Structure of the yeast CKM | | Descriptor: | Mediator of RNA polymerase II transcription subunit 12, Mediator of RNA polymerase II transcription subunit 13, Meiotic mRNA stability protein kinase SSN3, ... | | Authors: | Li, Y.C, Chao, T.C, Kim, H.J, Cholko, T, Chen, S.F, Nakanishi, K, Chang, C.E, Murakami, K, Garcia, B.A, Boyer, T.G, Tsai, K.L. | | Deposit date: | 2020-11-12 | | Release date: | 2021-01-27 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structure and noncanonical Cdk8 activation mechanism within an Argonaute-containing Mediator kinase module.
Sci Adv, 7, 2021
|
|
3MT1
 
 | | Crystal structure of putative carboxynorspermidine decarboxylase protein from Sinorhizobium meliloti | | Descriptor: | Putative carboxynorspermidine decarboxylase protein, SULFATE ION | | Authors: | Chang, C, Xu, X, Cui, H, Chin, S, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-04-29 | | Release date: | 2010-06-30 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of putative carboxynorspermidine decarboxylase protein from Sinorhizobium meliloti
To be Published
|
|
7KPV
 
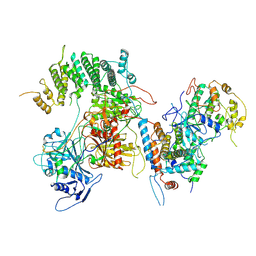 | | Structure of kinase and Central lobes of yeast CKM | | Descriptor: | Mediator of RNA polymerase II transcription subunit 12, Mediator of RNA polymerase II transcription subunit 13, Meiotic mRNA stability protein kinase SSN3, ... | | Authors: | Li, Y.C, Chao, T.C, Kim, H.J, Cholko, T, Chen, S.F, Nakanishi, K, Chang, C.E, Murakami, K, Garcia, B.A, Boyer, T.G, Tsai, K.L. | | Deposit date: | 2020-11-12 | | Release date: | 2021-01-27 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure and noncanonical Cdk8 activation mechanism within an Argonaute-containing Mediator kinase module.
Sci Adv, 7, 2021
|
|
3TKN
 
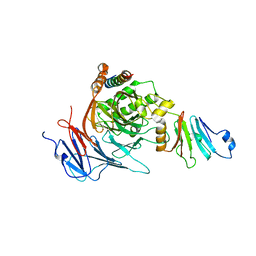 | | Structure of the Nup82-Nup159-Nup98 heterotrimer | | Descriptor: | Nucleoporin 98, Nucleoporin NUP159, Nucleoporin NUP82 | | Authors: | Stuwe, T.T, Hoelz, A. | | Deposit date: | 2011-08-28 | | Release date: | 2012-04-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Molecular basis for the anchoring of proto-oncoprotein nup98 to the cytoplasmic face of the nuclear pore complex.
J.Mol.Biol., 419, 2012
|
|
3K7Z
 
 | |
3M9Z
 
 | | Crystal Structure of extracellular domain of mouse NKR-P1A | | Descriptor: | Killer cell lectin-like receptor subfamily B member 1A, PHOSPHATE ION | | Authors: | Kolenko, P, Rozbesky, D, Bezouska, K, Hasek, J, Dohnalek, J. | | Deposit date: | 2010-03-23 | | Release date: | 2011-04-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular architecture of mouse activating NKR-P1 receptors.
J.Struct.Biol., 175, 2011
|
|
3V2H
 
 | | The crystal structure of D-beta-hydroxybutyrate dehydrogenase from Sinorhizobium meliloti | | Descriptor: | D-beta-hydroxybutyrate dehydrogenase | | Authors: | Zhang, Z, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-12-12 | | Release date: | 2011-12-28 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The crystal structure of D-beta-hydroxybutyrate dehydrogenase from Sinorhizobium meliloti
To be Published
|
|
7KFN
 
 | | Structure of Human Adenosine Deaminase Acting on dsRNA (ADAR2) bound to dsRNA containing a 2'-deoxy Benner's Base Z opposite the edited base | | Descriptor: | Double-stranded RNA-specific editase 1, Gli1 1W5 23mer RNA, Gli1 8AZ 23mer RNA, ... | | Authors: | Wilcox, X.E, Fisher, A.J, Beal, P.A. | | Deposit date: | 2020-10-14 | | Release date: | 2021-06-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Rational Design of RNA Editing Guide Strands: Cytidine Analogs at the Orphan Position.
J.Am.Chem.Soc., 143, 2021
|
|
3I5R
 
 | | PI3K SH3 domain in complex with a peptide ligand | | Descriptor: | Peptide ligand, Phosphatidylinositol 3-kinase regulatory subunit alpha | | Authors: | Batra-Safferling, R, Granzin, J, Modder, S, Hoffmann, S, Willbold, D. | | Deposit date: | 2009-07-06 | | Release date: | 2010-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural studies of the phosphatidylinositol 3-kinase (PI3K) SH3 domain in complex with a peptide ligand: role of the anchor residue in ligand binding.
Biol.Chem., 391, 2010
|
|
3I5S
 
 | | Crystal structure of PI3K SH3 | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, SULFATE ION | | Authors: | Batra-Safferling, R, Granzin, J, Modder, S, Hoffmann, S, Willbold, D. | | Deposit date: | 2009-07-06 | | Release date: | 2010-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural studies of the phosphatidylinositol 3-kinase (PI3K) SH3 domain in complex with a peptide ligand: role of the anchor residue in ligand binding.
Biol.Chem., 391, 2010
|
|
4OVV
 
 | | Crystal Structure of PI3Kalpha in complex with diC4-PIP2 | | Descriptor: | (2R)-3-{[(R)-HYDROXY{[(1R,2R,3S,4R,5R,6S)-2,3,6-TRIHYDROXY-4,5-BIS(PHOSPHONOOXY)CYCLOHEXYL]OXY}PHOSPHORYL]OXY}PROPANE-1 ,2-DIYL DIBUTANOATE, PHOSPHATE ION, Phosphatidylinositol 3-kinase regulatory subunit alpha, ... | | Authors: | Gabelli, S.B, Vogelstein, B, Miller, M, Amzel, L.M. | | Deposit date: | 2014-01-14 | | Release date: | 2014-09-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural basis of nSH2 regulation and lipid binding in PI3K alpha.
Oncotarget, 5, 2014
|
|
7LM2
 
 | | HUMAN PI3KDELTA IN COMPLEX WITH COMPOUND 3C | | Descriptor: | 1,2-ETHANEDIOL, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform, ... | | Authors: | Lesburg, C.A, Augustin, M. | | Deposit date: | 2021-02-05 | | Release date: | 2021-04-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Projected Dose Optimization of Amino- and Hydroxypyrrolidine Purine PI3K delta Immunomodulators.
J.Med.Chem., 64, 2021
|
|
3V8B
 
 | | Crystal Structure of a 3-ketoacyl-ACP reductase from Sinorhizobium meliloti 1021 | | Descriptor: | Putative dehydrogenase, possibly 3-oxoacyl-[acyl-carrier protein] reductase | | Authors: | Kumaran, D, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-12-22 | | Release date: | 2012-01-11 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of a 3-ketoacyl-ACP reductase from Sinorhizobium meliloti 1021
To be Published
|
|
3IAG
 
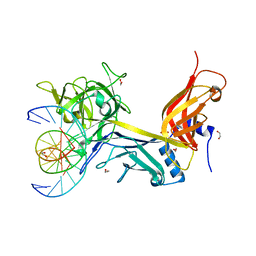 | | CSL (RBP-Jk) bound to HES-1 nonconsensus site | | Descriptor: | 1,2-ETHANEDIOL, 5'-D(*AP*AP*TP*CP*TP*TP*TP*CP*AP*CP*AP*CP*GP*AP*T)-3', 5'-D(*TP*TP*AP*TP*CP*GP*TP*GP*TP*GP*AP*AP*AP*GP*A)-3', ... | | Authors: | Friedmann, D.R, Kovall, R.A. | | Deposit date: | 2009-07-13 | | Release date: | 2009-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Thermodynamic and structural insights into CSL-DNA complexes.
Protein Sci., 19, 2010
|
|
4QKI
 
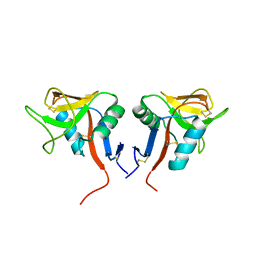 | | Dimeric form of human LLT1, a ligand for NKR-P1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C-type lectin domain family 2 member D | | Authors: | Skalova, T, Blaha, J, Harlos, K, Duskova, J, Koval, T, Stransky, J, Hasek, J, Vanek, O, Dohnalek, J. | | Deposit date: | 2014-06-06 | | Release date: | 2015-03-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Four crystal structures of human LLT1, a ligand of human NKR-P1, in varied glycosylation and oligomerization states
Acta Crystallogr.,Sect.D, 71, 2015
|
|
1DLW
 
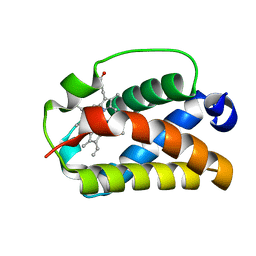 | | X-RAY CRYSTAL STRUCTURE OF TRUNCATED HEMOGLOBIN FROM P.CAUDATUM. | | Descriptor: | HEMOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Pesce, A, Couture, M, Guertin, M, Dewilde, S, Moens, L, Bolognesi, M. | | Deposit date: | 1999-12-13 | | Release date: | 2000-09-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | A novel two-over-two alpha-helical sandwich fold is characteristic of the truncated hemoglobin family.
EMBO J., 19, 2000
|
|
8GUB
 
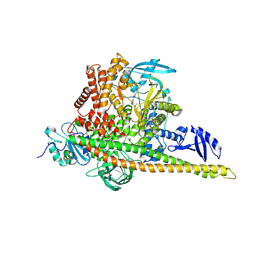 | | Cryo-EM structure of cancer-specific PI3Kalpha mutant H1047R in complex with BYL-719 | | Descriptor: | (2S)-N~1~-{4-methyl-5-[2-(1,1,1-trifluoro-2-methylpropan-2-yl)pyridin-4-yl]-1,3-thiazol-2-yl}pyrrolidine-1,2-dicarboxamide, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Liu, X, Zhou, Q, Hart, J.R, Xu, Y, Yang, S, Yang, D, Vogt, P.K, Wang, M.-W. | | Deposit date: | 2022-09-11 | | Release date: | 2022-11-23 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Cryo-EM structures of cancer-specific helical and kinase domain mutations of PI3K alpha.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
1D6E
 
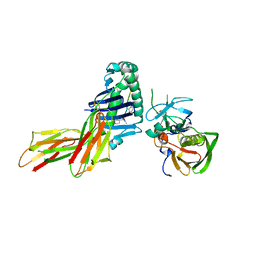 | | CRYSTAL STRUCTURE OF HLA-DR4 COMPLEX WITH PEPTIDOMIMETIC AND SEB | | Descriptor: | ENTEROTOXIN TYPE B, HLA CLASS II HISTOCOMPATIBILITY ANTIGEN, PEPTIDOMIMETIC INHIBITOR | | Authors: | Swain, A, Crowther, R, Kammlott, U. | | Deposit date: | 1999-10-13 | | Release date: | 2000-06-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Peptide and peptide mimetic inhibitors of antigen presentation by HLA-DR class II MHC molecules. Design, structure-activity relationships, and X-ray crystal structures.
J.Med.Chem., 43, 2000
|
|
1DLH
 
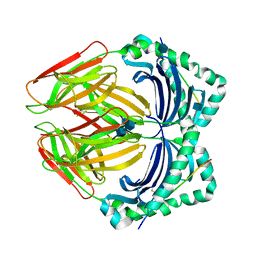 | |
8H36
 
 | | Cryo-EM Structure of the KBTBD2-CUL3-Rbx1-p85a dimeric complex | | Descriptor: | Cullin-3, E3 ubiquitin-protein ligase RBX1, Kelch repeat and BTB domain-containing protein 2, ... | | Authors: | Hu, Y, Mao, Q, Chen, Z, Sun, L. | | Deposit date: | 2022-10-08 | | Release date: | 2023-10-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Dynamic molecular architecture and substrate recruitment of cullin3-RING E3 ligase CRL3 KBTBD2.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8H37
 
 | | Cryo-EM Structure of the KBTBD2-CUL3-Rbx1-p85a tetrameric complex | | Descriptor: | Cullin-3, E3 ubiquitin-protein ligase RBX1, Kelch repeat and BTB domain-containing protein 2, ... | | Authors: | Hu, Y, Mao, Q, Chen, Z, Sun, L. | | Deposit date: | 2022-10-08 | | Release date: | 2023-10-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (7.52 Å) | | Cite: | Dynamic molecular architecture and substrate recruitment of cullin3-RING E3 ligase CRL3 KBTBD2.
Nat.Struct.Mol.Biol., 31, 2024
|
|
1D5M
 
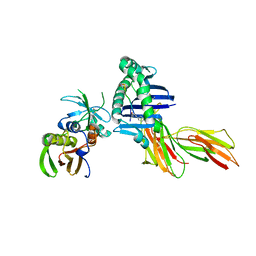 | | X-RAY CRYSTAL STRUCTURE OF HLA-DR4 COMPLEXED WITH PEPTIDE AND SEB | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ENTEROTOXIN TYPE B, HLA CLASS II HISTOCOMPATIBILITY ANTIGEN, ... | | Authors: | Swain, A.L, Crowther, R, Kammlott, U. | | Deposit date: | 1999-10-07 | | Release date: | 2000-06-28 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Peptide and peptide mimetic inhibitors of antigen presentation by HLA-DR class II MHC molecules. Design, structure-activity relationships, and X-ray crystal structures.
J.Med.Chem., 43, 2000
|
|
1DLY
 
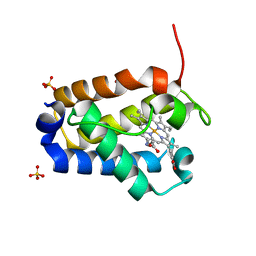 | | X-RAY CRYSTAL STRUCTURE OF HEMOGLOBIN FROM THE GREEN UNICELLULAR ALGA CHLAMYDOMONAS EUGAMETOS | | Descriptor: | 1,2-ETHANEDIOL, CYANIDE ION, HEMOGLOBIN, ... | | Authors: | Pesce, A, Couture, M, Guertin, M, Dewilde, S, Moens, L, Bolognesi, M. | | Deposit date: | 1999-12-13 | | Release date: | 2000-09-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A novel two-over-two alpha-helical sandwich fold is characteristic of the truncated hemoglobin family.
EMBO J., 19, 2000
|
|
1D5X
 
 | | X-RAY CRYSTAL STRUCTURE OF HLA-DR4 COMPLEXED WITH DIPEPTIDE MIMETIC AND SEB | | Descriptor: | DIPEPTIDE MIMETIC INHIBITOR, ENTEROTOXIN TYPE B, HLA CLASS II HISTOCOMPATIBILITY ANTIGEN | | Authors: | Swain, A, Crowther, R, Kammlott, U. | | Deposit date: | 1999-10-12 | | Release date: | 2000-06-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Peptide and peptide mimetic inhibitors of antigen presentation by HLA-DR class II MHC molecules. Design, structure-activity relationships, and X-ray crystal structures.
J.Med.Chem., 43, 2000
|
|
