1JII
 
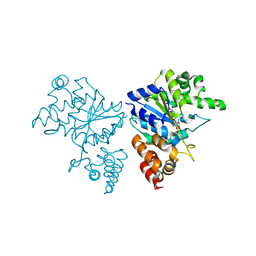 | | Crystal structure of S. aureus TyrRS in complex with SB-219383 | | Descriptor: | [2-AMINO-3-(4-HYDROXY-PHENYL)-PROPIONYLAMINO]- (2,4,5,8-TETRAHYDROXY-7-OXA-2-AZA-BICYCLO[3.2.1]OCT-3-YL)- ACETIC ACID, tyrosyl-tRNA synthetase | | Authors: | Qiu, X, Janson, C.A, Smith, W.W, Jarvest, R.L. | | Deposit date: | 2001-07-02 | | Release date: | 2001-10-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of Staphylococcus aureus tyrosyl-tRNA synthetase in complex with a class of potent and specific inhibitors.
Protein Sci., 10, 2001
|
|
8A8Z
 
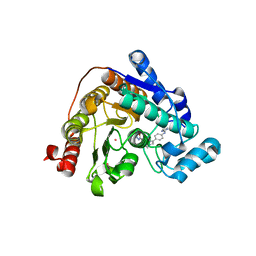 | | Crystal structure of Danio rerio HDAC6 CD2 in complex with in situ enzymatically hydrolyzed DFMO-based ITF5924 | | Descriptor: | 4-[[4-[4-(imidazolidin-2-ylideneamino)phenyl]-1,2,3-triazol-1-yl]methyl]benzohydrazide, Histone deacetylase 6, POTASSIUM ION, ... | | Authors: | Zrubek, K, Sandrone, G, Cukier, C.D, Stevenazzi, A. | | Deposit date: | 2022-06-27 | | Release date: | 2023-01-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Difluoromethyl-1,3,4-oxadiazoles are slow-binding substrate analog inhibitors of histone deacetylase 6 with unprecedented isotype selectivity.
J.Biol.Chem., 299, 2022
|
|
6E6J
 
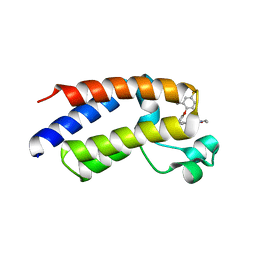 | | BRD2_Bromodomain2 complex with inhibitor 744 | | Descriptor: | Bromodomain-containing protein 2, N-ethyl-4-[2-(4-fluoro-2,6-dimethylphenoxy)-5-(2-hydroxypropan-2-yl)phenyl]-6-methyl-7-oxo-6,7-dihydro-1H-pyrrolo[2,3-c]pyridine-2-carboxamide | | Authors: | Longenecker, K.L, Park, C.H, Bigelow, L. | | Deposit date: | 2018-07-25 | | Release date: | 2019-07-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Selective inhibition of the BD2 bromodomain of BET proteins in prostate cancer.
Nature, 578, 2020
|
|
1JIK
 
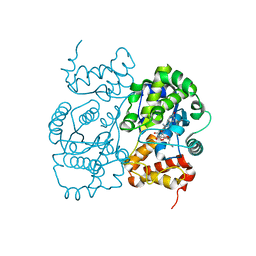 | | Crystal structure of S. aureus TyrRS in complex with SB-243545 | | Descriptor: | [2-AMINO-3-(4-HYDROXY-PHENYL)-PROPIONYLAMINO]-(1,3,4,5-TETRAHYDROXY-4-HYDROXYMETHYL-PIPERIDIN-2-YL)- ACETIC ACID BUTYL ESTER, tyrosyl-tRNA synthetase | | Authors: | Qiu, X, Janson, C.A, Smith, W.W, Jarvest, R.L. | | Deposit date: | 2001-07-02 | | Release date: | 2001-10-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of Staphylococcus aureus tyrosyl-tRNA synthetase in complex with a class of potent and specific inhibitors.
Protein Sci., 10, 2001
|
|
7KL0
 
 | | Cocrystal structure of human CaMKII-alpha (CAMK2A)kinase domain and GluN2B(S1303D) | | Descriptor: | 1,2-ETHANEDIOL, Calcium/calmodulin-dependent protein kinase type II subunit alpha, Glutamate receptor ionotropic, ... | | Authors: | Ozden, C, Stratton, M.M, Garman, S.C. | | Deposit date: | 2020-10-28 | | Release date: | 2020-12-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | CaMKII binds both substrates and activators at the active site.
Cell Rep, 40, 2022
|
|
1LCB
 
 | |
7L05
 
 | | Complex of novel maytansinoid M24 bound to T2R-TTL (two tubulin alpha/beta heterodimers, RB3 stathmin-like domain, and tubulin tyrosine ligase) | | Descriptor: | (1S,2R,3S,5S,6S,16E,18E,20R,21S)-11-chloro-21-hydroxy-12,20-dimethoxy-2,5,9,16-tetramethyl-8,23-dioxo-4,24-dioxa-9,22-diazatetracyclo[19.3.1.1~10,14~.0~3,5~]hexacosa-10(26),11,13,16,18-pentaen-6-yl (2S)-2-{methyl[3-(methylamino)propanoyl]amino}propanoate (non-preferred name), 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Franklin, M.C. | | Deposit date: | 2020-12-11 | | Release date: | 2021-12-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | A Biparatopic Antibody-Drug Conjugate to Treat MET-Expressing Cancers, Including Those that Are Unresponsive to MET Pathway Blockade.
Mol.Cancer Ther., 20, 2021
|
|
7L5A
 
 | | Crystal structure of the photosensory module from Xanthomonas campestris bacteriophytochrome XccBphP in the Pfr state | | Descriptor: | BILIVERDINE IX ALPHA, Bacteriophytochrome | | Authors: | Otero, L.H, Antelo, G, Sanchez-Lamas, M, Klinke, S, Goldbaum, F.A, Rinaldi, J, Bonomi, H.R. | | Deposit date: | 2020-12-21 | | Release date: | 2021-12-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural basis for the Pr-Pfr long-range signaling mechanism of a full-length bacterial phytochrome at the atomic level.
Sci Adv, 7, 2021
|
|
7L59
 
 | | Crystal structure of the dark-adapted full-length bacteriophytochrome XccBphP-G454E variant from Xanthomonas campestris in the Pfr state | | Descriptor: | BILIVERDINE IX ALPHA, Bacteriophytochrome | | Authors: | Otero, L.H, Antelo, G, Sanchez-Lamas, M, Klinke, S, Goldbaum, F.A, Rinaldi, J, Bonomi, H.R. | | Deposit date: | 2020-12-21 | | Release date: | 2021-12-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Structural basis for the Pr-Pfr long-range signaling mechanism of a full-length bacterial phytochrome at the atomic level.
Sci Adv, 7, 2021
|
|
7LTJ
 
 | | Room-temperature X-ray structure of SARS-CoV-2 main protease (3CL Mpro) in complex with a non-covalent inhibitor Mcule-5948770040 | | Descriptor: | 3C-like proteinase, 6-[4-(3,4-dichlorophenyl)piperazin-1-yl]carbonyl-1~{H}-pyrimidine-2,4-dione | | Authors: | Kovalevsky, A, Kneller, D.W, Coates, L. | | Deposit date: | 2021-02-19 | | Release date: | 2021-03-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High-Throughput Virtual Screening and Validation of a SARS-CoV-2 Main Protease Noncovalent Inhibitor.
J.Chem.Inf.Model., 62, 2022
|
|
1LCA
 
 | | LACTOBACILLUS CASEI THYMIDYLATE SYNTHASE TERNARY COMPLEX WITH DUMP AND CB3717 | | Descriptor: | 10-PROPARGYL-5,8-DIDEAZAFOLIC ACID, 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, THYMIDYLATE SYNTHASE | | Authors: | Birdsall, D.L, Finer-Moore, J, Stroud, R.M. | | Deposit date: | 1995-06-22 | | Release date: | 1995-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Refined structures of substrate-bound and phosphate-bound thymidylate synthase from Lactobacillus casei.
J.Mol.Biol., 232, 1993
|
|
6PL0
 
 | | Crystal structure of the dark-adapted full-length bacteriophytochrome XccBphP from Xanthomonas campestris in the Pr state bound to BV chromophore | | Descriptor: | BILIVERDINE IX ALPHA, Bacteriophytochrome | | Authors: | Otero, L.H, Sirigu, S, Klinke, S, Goldbaum, F, Chavas, L, Rinaldi, J, Bonomi, H.R. | | Deposit date: | 2019-06-30 | | Release date: | 2020-12-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Structural basis for the Pr-Pfr long-range signaling mechanism of a full-length bacterial phytochrome at the atomic level.
Sci Adv, 7, 2021
|
|
7RFY
 
 | | Importin alpha3 in complex with MERS ORF4B | | Descriptor: | Importin subunit alpha-3, ORF4b | | Authors: | Munasinghe, T.S, Tsimbalyuk, S, Roby, J.A, Aragao, D, Forwood, J.K. | | Deposit date: | 2021-07-14 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | MERS-CoV ORF4b employs an unusual binding mechanism to target IMP alpha and block innate immunity.
Nat Commun, 13, 2022
|
|
7RG0
 
 | | Importin alpha2 in complex with MERS ORF4B R24A mutant | | Descriptor: | Importin subunit alpha-1, ORF4b | | Authors: | Munasinghe, T.S, Tsimbalyuk, S, Roby, J.A, Aragao, D, Forwood, J.K. | | Deposit date: | 2021-07-14 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | MERS-CoV ORF4b employs an unusual binding mechanism to target IMP alpha and block innate immunity.
Nat Commun, 13, 2022
|
|
7RFX
 
 | | Importin alpha2 in complex with MERS ORF4B | | Descriptor: | Importin subunit alpha-1, ORF4b | | Authors: | Munasinghe, T.S, Tsimbalyuk, S, Roby, J.A, Aragao, D, Forwood, J.K. | | Deposit date: | 2021-07-14 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | MERS-CoV ORF4b employs an unusual binding mechanism to target IMP alpha and block innate immunity.
Nat Commun, 13, 2022
|
|
5IKN
 
 | |
7RFZ
 
 | | Importin alpha 2 in complex with MERS ORF4B NLS peptide | | Descriptor: | Importin subunit alpha-1, ORF4b | | Authors: | Munasinghe, T.S, Tsimbalyuk, S, Roby, J.A, Aragao, D, Forwood, J.K. | | Deposit date: | 2021-07-14 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | MERS-CoV ORF4b employs an unusual binding mechanism to target IMP alpha and block innate immunity.
Nat Commun, 13, 2022
|
|
7RG4
 
 | | Importin alpha2 in complex with p50 NLS | | Descriptor: | Importin subunit alpha-1, Isoform 3 of Nuclear factor NF-kappa-B p105 subunit | | Authors: | Smith, K.M, Tsimbalyuk, S, Aragao, D, Forwood, J.K. | | Deposit date: | 2021-07-14 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | MERS-CoV ORF4b employs an unusual binding mechanism to target IMP alpha and block innate immunity.
Nat Commun, 13, 2022
|
|
7RG2
 
 | | Importin alpha2 in complex with MERS ORF4B R33A mutant | | Descriptor: | Importin subunit alpha-1, ORF4b | | Authors: | Munasinghe, T.S, Tsimbalyuk, S, Roby, J.A, Aragao, D, Forwood, J.K. | | Deposit date: | 2021-07-14 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | MERS-CoV ORF4b employs an unusual binding mechanism to target IMP alpha and block innate immunity.
Nat Commun, 13, 2022
|
|
7RG5
 
 | | Importin alpha3 in complex with p50 NLS | | Descriptor: | Importin subunit alpha-3, Isoform 3 of Nuclear factor NF-kappa-B p105 subunit | | Authors: | Smith, K.M, Tsimbalyuk, S, Aragao, D, Forwood, J.K. | | Deposit date: | 2021-07-14 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | MERS-CoV ORF4b employs an unusual binding mechanism to target IMP alpha and block innate immunity.
Nat Commun, 13, 2022
|
|
7RG3
 
 | | Importin alpha2 in complex with MERS ORF4B R37A mutant | | Descriptor: | Importin subunit alpha-1, ORF4b | | Authors: | Munasinghe, T.S, Tsimbalyuk, S, Roby, J.A, Aragao, D, Forwood, J.K. | | Deposit date: | 2021-07-14 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | MERS-CoV ORF4b employs an unusual binding mechanism to target IMP alpha and block innate immunity.
Nat Commun, 13, 2022
|
|
1LPG
 
 | | CRYSTAL STRUCTURE OF FXA IN COMPLEX WITH 79. | | Descriptor: | Blood coagulation factor Xa, CALCIUM ION, [4-({[5-BENZYLOXY-1-(3-CARBAMIMIDOYL-BENZYL)-1H-INDOLE-2-CARBONYL]-AMINO}-METHYL)-PHENYL]-TRIMETHYL-AMMONIUM | | Authors: | Schreuder, H.A, Brachvogel, V, Liesum, A. | | Deposit date: | 2002-05-08 | | Release date: | 2003-05-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design and Quantitative Structure-Activity relationship of 3-amidinobenzyl-1H-indole-2-carboxamides as potent, nonchiral, and selective inhibitors of blood coagulation factor Xa.
J.Med.Chem., 45, 2002
|
|
1LPZ
 
 | | CRYSTAL STRUCTURE OF FXA IN COMPLEX WITH 41. | | Descriptor: | 1-(3-carbamimidoylbenzyl)-N-(3,5-dichlorobenzyl)-4-methyl-1H-indole-2-carboxamide, Blood coagulation factor Xa, CALCIUM ION | | Authors: | Schreuder, H.A, Brachvogel, V, Liesum, A. | | Deposit date: | 2002-05-08 | | Release date: | 2003-05-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Design and Quantitative Structure-Activity relationship of 3-amidinobenzyl-1H-indole-2-carboxamides as potent, nonchiral, and selective inhibitors of blood coagulation factor Xa
J.Biol.Chem., 45, 2002
|
|
1LQE
 
 | | CRYSTAL STRUCTURE OF TRYPSIN IN COMPLEX WITH 79. | | Descriptor: | CALCIUM ION, SULFATE ION, TRYPSIN, ... | | Authors: | Schreuder, H.A, Liesum, A. | | Deposit date: | 2002-05-10 | | Release date: | 2003-05-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Design and quantitative structure-activity relationship of 3-amidinobenzyl-1H-indole-2-carboxamides
as potent, nonchiral, and selective inhibitors of blood coagulation factor Xa.
J.Med.Chem., 45, 2002
|
|
7MMO
 
 | | LY-CoV1404 neutralizing antibody against SARS-CoV-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, LY-CoV1404 Fab heavy chain, LY-CoV1404 Fab light chain, ... | | Authors: | Hendle, J, Pustilnik, A, Sauder, J.M, Coleman, K.A, Boyles, J.S, Dickinson, C.D. | | Deposit date: | 2021-04-30 | | Release date: | 2021-05-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.427 Å) | | Cite: | LY-CoV1404 (bebtelovimab) potently neutralizes SARS-CoV-2 variants.
Biorxiv, 2022
|
|
