8DYQ
 
 | | Crystal structure of Neisseria gonorrhoeae carbonic anhydrase with Acetazolamide | | Descriptor: | 5-ACETAMIDO-1,3,4-THIADIAZOLE-2-SULFONAMIDE, Carbonic anhydrase, SULFATE ION, ... | | Authors: | Marapaka, A.K, Das, C, Flaherty, D.P, Yadav, R. | | Deposit date: | 2022-08-04 | | Release date: | 2022-12-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural Characterization of Thiadiazolesulfonamide Inhibitors Bound to Neisseria gonorrhoeae alpha-Carbonic Anhydrase.
Acs Med.Chem.Lett., 14, 2023
|
|
7ER3
 
 | | Crystal structure of beta-lactoglobulin complexed with chloroquine | | Descriptor: | (4S)-N~4~-(7-chloroquinolin-4-yl)-N~1~,N~1~-diethylpentane-1,4-diamine, Major allergen beta-lactoglobulin | | Authors: | Yao, Q, Ma, J, Xing, Y, Zang, J. | | Deposit date: | 2021-05-05 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.598 Å) | | Cite: | Binding of Chloroquine to Whey Protein Relieves Its Cytotoxicity while Enhancing Its Uptake by Cells.
J.Agric.Food Chem., 69, 2021
|
|
8OK0
 
 | | Crystal structure of human NQO1 in complex with the inhibitor PMSF | | Descriptor: | ACETYL GROUP, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Martin-Garcia, J.M, Grieco, A, Ruiz-Fresneda, M.A. | | Deposit date: | 2023-03-26 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural dynamics at the active site of the cancer-associated flavoenzyme NQO1 probed by chemical modification with PMSF.
Febs Lett., 597, 2023
|
|
8DR2
 
 | | Crystal structure of Neisseria gonorrhoeae carbonic anhydrase with 2-cyclohexyl-N-(5-sulfamoyl-1,3,4-thiadiazol-2-yl)acetamide | | Descriptor: | 2-cyclohexyl-N-(5-sulfamoyl-1,3,4-thiadiazol-2-yl)acetamide, Carbonic anhydrase, SULFATE ION, ... | | Authors: | Marapaka, A.K, Das, C, Flaherty, D.P, Yadav, R. | | Deposit date: | 2022-07-20 | | Release date: | 2022-12-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Structural Characterization of Thiadiazolesulfonamide Inhibitors Bound to Neisseria gonorrhoeae alpha-Carbonic Anhydrase.
Acs Med.Chem.Lett., 14, 2023
|
|
6N5F
 
 | | Crystal structure of an epoxide hydrolase from Trichoderma reesei in complex with inhibitor 3 | | Descriptor: | Epoxide hydrolase TrEH, N-(8-amino-8-oxooctyl)nonanamide | | Authors: | Oliveira, G.S, Adriani, P.P, Ribeiro, J.A, Morisseau, C, Hammock, B.D, Dias, M.V, Chambergo, F.S. | | Deposit date: | 2018-11-21 | | Release date: | 2019-11-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | The molecular structure of an epoxide hydrolase from Trichoderma reesei in complex with urea or amide-based inhibitors.
Int. J. Biol. Macromol., 129, 2019
|
|
6N5G
 
 | | Crystal structure of an epoxide hydrolase from Trichoderma reesei in complex with inhibitor 2 | | Descriptor: | 4-[(quinolin-3-yl)methyl]-N-[4-(trifluoromethoxy)phenyl]piperidine-1-carboxamide, Epoxide hydrolase TrEH | | Authors: | Oliveira, G.S, Adriani, P.P, Ribeiro, J.A, Morisseau, C, Hammock, B.D, Dias, M.V, Chambergo, F.S. | | Deposit date: | 2018-11-21 | | Release date: | 2019-11-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The molecular structure of an epoxide hydrolase from Trichoderma reesei in complex with urea or amide-based inhibitors.
Int. J. Biol. Macromol., 129, 2019
|
|
8DRB
 
 | | Crystal structure of Neisseria gonorrhoeae carbonic anhydrase with 3-phenyl-N-(5-sulfamoyl-1,3,4-thiadiazol-2-yl)propanamide | | Descriptor: | 3-phenyl-N-(5-sulfamoyl-1,3,4-thiadiazol-2-yl)propanamide, Carbonic anhydrase, SULFATE ION, ... | | Authors: | Marapaka, A.K, Das, C, Flaherty, D.P, Yadav, R. | | Deposit date: | 2022-07-20 | | Release date: | 2022-12-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structural Characterization of Thiadiazolesulfonamide Inhibitors Bound to Neisseria gonorrhoeae alpha-Carbonic Anhydrase.
Acs Med.Chem.Lett., 14, 2023
|
|
4QU3
 
 | | GES-2 ertapenem acyl-enzyme complex | | Descriptor: | (4R,5S)-3-({(3S,5S)-5-[(3-carboxyphenyl)carbamoyl]pyrrolidin-3-yl}sulfanyl)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-4-methyl-4,5-dihydro-1H-pyrrole-2-carboxylic acid, 1,2-ETHANEDIOL, Beta-lactamase GES-2, ... | | Authors: | Stewart, N.K, Smith, C.A, Frase, H, Black, D.J, Vakulenko, S.B. | | Deposit date: | 2014-07-10 | | Release date: | 2014-12-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.402 Å) | | Cite: | Kinetic and Structural Requirements for Carbapenemase Activity in GES-Type beta-Lactamases.
Biochemistry, 54, 2015
|
|
3B9O
 
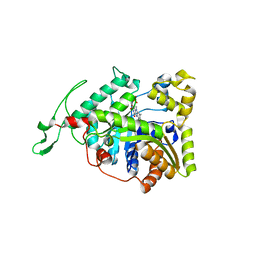 | | long-chain alkane monooxygenase (LadA) in complex with coenzyme FMN | | Descriptor: | Alkane monooxygenase, FLAVIN MONONUCLEOTIDE | | Authors: | Li, L, Yang, W, Xu, F, Bartlam, M, Rao, Z. | | Deposit date: | 2007-11-06 | | Release date: | 2008-01-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of long-chain alkane monooxygenase (LadA) in complex with coenzyme FMN: unveiling the long-chain alkane hydroxylase
J.Mol.Biol., 376, 2008
|
|
4R5B
 
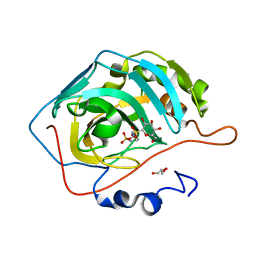 | |
1YKW
 
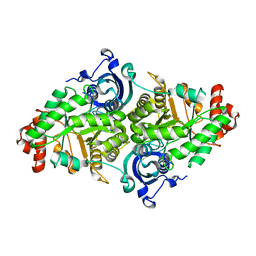 | |
8DPC
 
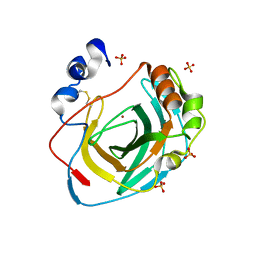 | | Crystal structure of carbonic anhydrase from Neisseria gonorrhoeae | | Descriptor: | Carbonic anhydrase, SULFATE ION, ZINC ION | | Authors: | Marapaka, A.K, Das, C, Flaherty, D.P, Yadav, R. | | Deposit date: | 2022-07-15 | | Release date: | 2022-12-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structural Characterization of Thiadiazolesulfonamide Inhibitors Bound to Neisseria gonorrhoeae alpha-Carbonic Anhydrase.
Acs Med.Chem.Lett., 14, 2023
|
|
1YLU
 
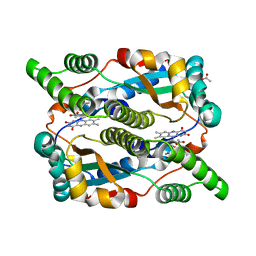 | | The structure of E. coli nitroreductase with bound acetate, crystal form 2 | | Descriptor: | ACETATE ION, FLAVIN MONONUCLEOTIDE, Oxygen-insensitive NAD(P)H nitroreductase | | Authors: | Race, P.R, Lovering, A.L, Green, R.M, Ossor, A, White, S.A, Searle, P.F, Wrighton, C.J, Hyde, E.I. | | Deposit date: | 2005-01-19 | | Release date: | 2005-02-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and mechanistic studies of Escherichia coli nitroreductase with the antibiotic nitrofurazone. Reversed binding orientations in different redox states of the enzyme.
J.Biol.Chem., 280, 2005
|
|
7LOT
 
 | |
8K5C
 
 | |
8OL6
 
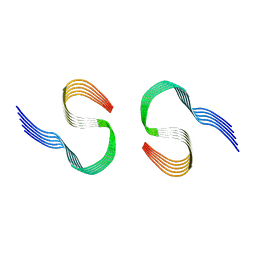 | | Murine type II Abeta fibril from tgAPPSwe mouse | | Descriptor: | Amyloid-beta protein 42 | | Authors: | Zielinski, M, Peralta Reyes, F.S, Gremer, L, Schemmert, S, Frieg, B, Willuweit, A, Donner, L, Elvers, M, Nilsson, L.N.G, Syvanen, S, Sehlin, D, Ingelsson, M, Willbold, D, Schroeder, G.F. | | Deposit date: | 2023-03-30 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM of A beta fibrils from mouse models find tg-APP ArcSwe fibrils resemble those found in patients with sporadic Alzheimer's disease.
Nat.Neurosci., 26, 2023
|
|
8K5D
 
 | |
8K5B
 
 | |
8K2X
 
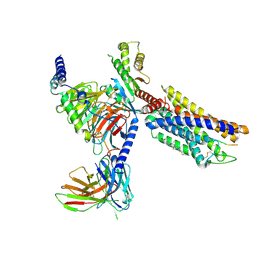 | | CXCR3-DNGi complex activated by CXCL10 | | Descriptor: | C-X-C motif chemokine 10, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Jiao, H.Z, Hu, H.L. | | Deposit date: | 2023-07-14 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure basis for the modulation of CXC chemokine receptor 3 by antagonist AMG487.
Cell Discov, 9, 2023
|
|
8OLN
 
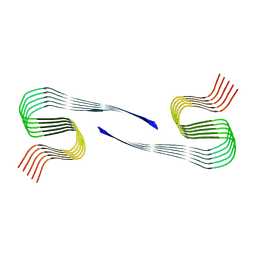 | | DI1 Abeta fibril from tg-SwDI mouse | | Descriptor: | Amyloid-beta protein 42 | | Authors: | Zielinski, M, Peralta Reyes, F.S, Gremer, L, Schemmert, S, Frieg, B, Willuweit, A, Donner, L, Elvers, M, Nilsson, L.N.G, Syvanen, S, Sehlin, D, Ingelsson, M, Willbold, D, Schroeder, G.F. | | Deposit date: | 2023-03-30 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM of A beta fibrils from mouse models find tg-APP ArcSwe fibrils resemble those found in patients with sporadic Alzheimer's disease.
Nat.Neurosci., 26, 2023
|
|
8OLQ
 
 | | DI3 Abeta fibril from tg-SwDI mouse | | Descriptor: | Amyloid-beta protein 42 | | Authors: | Zielinski, M, Peralta Reyes, F.S, Gremer, L, Schemmert, S, Frieg, B, Willuweit, A, Donner, L, Elvers, M, Nilsson, L.N.G, Syvanen, S, Sehlin, D, Ingelsson, M, Willbold, D, Schroeder, G.F. | | Deposit date: | 2023-03-30 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM of A beta fibrils from mouse models find tg-APP ArcSwe fibrils resemble those found in patients with sporadic Alzheimer's disease.
Nat.Neurosci., 26, 2023
|
|
4QSA
 
 | | Crystal structure of human carbonic anhydrase isozyme II with 2-chloro-4-{[(4-methyl-6-oxo-1,6-dihydropyrimidin-2-yl)thio]acetyl}benzenesulfonamide | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-chloro-4-{[(4-methyl-6-oxo-1,6-dihydropyrimidin-2-yl)sulfanyl]acetyl}benzenesulfonamide, Carbonic anhydrase 2, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2014-07-03 | | Release date: | 2015-01-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Intrinsic Thermodynamics and Structure Correlation of Benzenesulfonamides with a Pyrimidine Moiety Binding to Carbonic Anhydrases I, II, VII, XII, and XIII
Plos One, 9, 2014
|
|
8OL3
 
 | | Murine type III Abeta fibril from APP/PS1 mouse | | Descriptor: | Amyloid-beta protein 42 | | Authors: | Zielinski, M, Peralta Reyes, F.S, Gremer, L, Schemmert, S, Frieg, B, Willuweit, A, Donner, L, Elvers, M, Nilsson, L.N.G, Syvanen, S, Sehlin, D, Ingelsson, M, Willbold, D, Schroeder, G.F. | | Deposit date: | 2023-03-30 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM of A beta fibrils from mouse models find tg-APP ArcSwe fibrils resemble those found in patients with sporadic Alzheimer's disease.
Nat.Neurosci., 26, 2023
|
|
8OL2
 
 | | Murine type II Abeta fibril from APP23 mouse | | Descriptor: | Amyloid-beta protein 42 | | Authors: | Zielinski, M, Peralta Reyes, F.S, Gremer, L, Schemmert, S, Frieg, B, Willuweit, A, Donner, L, Elvers, M, Nilsson, L.N.G, Syvanen, S, Sehlin, D, Ingelsson, M, Willbold, D, Schroeder, G.F. | | Deposit date: | 2023-03-30 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM of A beta fibrils from mouse models find tg-APP ArcSwe fibrils resemble those found in patients with sporadic Alzheimer's disease.
Nat.Neurosci., 26, 2023
|
|
8OL5
 
 | | Murine type II Abeta fibril from ARTE10 mouse | | Descriptor: | Amyloid-beta protein 42 | | Authors: | Zielinski, M, Peralta Reyes, F.S, Gremer, L, Schemmert, S, Frieg, B, Willuweit, A, Donner, L, Elvers, M, Nilsson, L.N.G, Syvanen, S, Sehlin, D, Ingelsson, M, Willbold, D, Schroeder, G.F. | | Deposit date: | 2023-03-30 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM of A beta fibrils from mouse models find tg-APP ArcSwe fibrils resemble those found in patients with sporadic Alzheimer's disease.
Nat.Neurosci., 26, 2023
|
|
