5PO7
 
 | | PanDDA analysis group deposition -- Crystal Structure of BRD1 in complex with N11083a | | Descriptor: | 1,2-ETHANEDIOL, 1,3-dimethyl-5-(methylamino)-6-nitro-1,3-dihydro-2H-benzimidazol-2-one, Bromodomain-containing protein 1, ... | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
6DRT
 
 | |
4ZXH
 
 | | Crystal Structure of holo-AB3403 a four domain nonribosomal peptide synthetase from Acinetobacter Baumanii | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 4'-PHOSPHOPANTETHEINE, ABBFA_003403, ... | | Authors: | Drake, E.J, Miller, B.R, Allen, C.L, Gulick, A.M. | | Deposit date: | 2015-05-20 | | Release date: | 2015-12-30 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of two distinct conformations of holo-non-ribosomal peptide synthetases.
Nature, 529, 2016
|
|
7K6E
 
 | | SARS-CoV-2 Main Protease Co-Crystal Structure with Telaprevir Determined from Crystals Grown with 40 nL Acoustically Ejected Mpro Droplets at 1.63 A Resolution (Direct Vitrification) | | Descriptor: | (1S,3aR,6aS)-2-[(2S)-2-({(2S)-2-cyclohexyl-2-[(pyrazin-2-ylcarbonyl)amino]acetyl}amino)-3,3-dimethylbutanoyl]-N-[(2R,3S)-1-(cyclopropylamino)-2-hydroxy-1-oxohexan-3-yl]octahydrocyclopenta[c]pyrrole-1-carboxamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Kreitler, D.F, Andi, B, Kumaran, D, Soares, A.S, Shi, W, Jakoncic, J, Fuchs, M.R, Keereetaweep, J, Shanklin, J, McSweeney, S. | | Deposit date: | 2020-09-19 | | Release date: | 2020-09-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Hepatitis C virus NS3/4A inhibitors and other drug-like compounds as covalent binders of SARS-CoV-2 main protease.
Sci Rep, 12, 2022
|
|
3G5W
 
 | | Crystal structure of Blue Copper Oxidase from Nitrosomonas europaea | | Descriptor: | COPPER (II) ION, CU-O LINKAGE, CU-O-CU LINKAGE, ... | | Authors: | Lawton, T.J, Sayavedra-Soto, L.A, Arp, D.J, Rosenzweig, A.C. | | Deposit date: | 2009-02-05 | | Release date: | 2009-02-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a two-domain multicopper oxidase: implications for the evolution of multicopper blue proteins.
J.Biol.Chem., 284, 2009
|
|
6LPV
 
 | |
4QY7
 
 | |
3KMN
 
 | | Crystal Structure of the Human Apo GST Pi C47S/Y108V Double Mutant | | Descriptor: | CALCIUM ION, CARBONATE ION, Glutathione S-transferase P, ... | | Authors: | Parker, L.J. | | Deposit date: | 2009-11-11 | | Release date: | 2010-03-23 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Diuretic drug binding to human glutathione transferase P1-1: potential role of CYS101 revealed in the double mutant C47S/Y108V
to be published
|
|
2XSG
 
 | | Structure of the gh92 family glycosyl hydrolase ccman5 | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, CCMAN5, ... | | Authors: | Baranova, E, Tiels, P, Remaut, H. | | Deposit date: | 2010-09-29 | | Release date: | 2011-10-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Bacterial Glycosidase Enables Mannose-6-Phosphate Modification and Improved Cellular Uptake of Yeast-Produced Recombinant Human Lysosomal Enzymes.
Nat.Biotechnol., 30, 2012
|
|
5QSW
 
 | | PanDDA analysis group deposition -- Crystal Structure of human STAG1 in complex with Z2856434884 | | Descriptor: | 1-[4-(3-phenylpropyl)piperazin-1-yl]ethan-1-one, Cohesin subunit SA-1 | | Authors: | Newman, J.A, Katis, V.L, Gavard, A.E, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2019-05-25 | | Release date: | 2019-08-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5T6Z
 
 | | KIR3DL1 in complex with HLA-B*57:01-TW10 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, Decapeptide: THR-SER-THR-LEU-GLN-GLU-GLN-ILE-GLY-TRP, ... | | Authors: | Pymm, P, Rossjohn, J, Vivian, J.P. | | Deposit date: | 2016-09-02 | | Release date: | 2017-03-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | MHC-I peptides get out of the groove and enable a novel mechanism of HIV-1 escape.
Nat. Struct. Mol. Biol., 24, 2017
|
|
6M69
 
 | | Crystal structure of Mycobacterium smegmatis MutT1 in complex with GMPPCP (GDP) | | Descriptor: | 1,2-ETHANEDIOL, GUANOSINE-5'-DIPHOSPHATE, Hydrolase, ... | | Authors: | Raj, P, Karthik, S, Arif, S.M, Varshney, U, Vijayan, M. | | Deposit date: | 2020-03-13 | | Release date: | 2020-10-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Plasticity, ligand conformation and enzyme action of Mycobacterium smegmatis MutT1.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
3G83
 
 | |
6M77
 
 | |
4R6R
 
 | | Jacalin-carbohydrate interactions. Distortion of the ligand as a determinant of affinity. | | Descriptor: | 1,2-ETHANEDIOL, 4-nitrophenyl beta-D-galactopyranoside, Agglutinin alpha chain, ... | | Authors: | Abhinav, K.V, Sharma, K, Swaminathan, C.P, Surolia, A, Vijayan, M. | | Deposit date: | 2014-08-26 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Jacalin-carbohydrate interactions: distortion of the ligand molecule as a determinant of affinity.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3G7F
 
 | | Crystal structure of Blastochloris viridis heterodimer mutant reaction center | | Descriptor: | 15-cis-1,2-dihydroneurosporene, BACTERIOCHLOROPHYLL B, BACTERIOPHEOPHYTIN B, ... | | Authors: | Ponomarenko, N.S, Li, L, Tereshko, V, Ismagilov, R.F, Norris Jr, J.R. | | Deposit date: | 2009-02-09 | | Release date: | 2009-09-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and spectropotentiometric analysis of Blastochloris viridis heterodimer mutant reaction center
Biochim.Biophys.Acta, 1788, 2009
|
|
4KIO
 
 | | Kinase domain mutant of human Itk in complex with a covalently-binding inhibitor | | Descriptor: | 1-[(3S)-3-{[4-(morpholin-4-ylmethyl)-6-([1,3]thiazolo[5,4-b]pyridin-2-ylamino)pyrimidin-2-yl]amino}pyrrolidin-1-yl]prop-2-en-1-one, 1-[(3S)-3-{[4-(morpholin-4-ylmethyl)-6-([1,3]thiazolo[5,4-b]pyridin-2-ylamino)pyrimidin-2-yl]amino}pyrrolidin-1-yl]propan-1-one, GLYCEROL, ... | | Authors: | Somers, D.O. | | Deposit date: | 2013-05-02 | | Release date: | 2013-08-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Discovery of novel irreversible inhibitors of interleukin (IL)-2-inducible tyrosine kinase (Itk) by targeting cysteine 442 in the ATP pocket.
J.Biol.Chem., 288, 2013
|
|
5YOV
 
 | | Crystal structure of BRD4-BD1 bound with hjp126 | | Descriptor: | (3~{R})-4-cyclopentyl-~{N}-(2,4-dimethylphenyl)-1,3-dimethyl-2-oxidanylidene-3~{H}-quinoxaline-6-carboxamide, Bromodomain-containing protein 4 | | Authors: | Xiong, B, Hu, J, Li, Y, Cao, D. | | Deposit date: | 2017-10-31 | | Release date: | 2018-11-07 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.451 Å) | | Cite: | Structure-based optimization of a series of selective BET inhibitors containing aniline or indoline groups.
Eur.J.Med.Chem., 150, 2018
|
|
3G8I
 
 | | Aleglitazar, a new, potent, and balanced PPAR alpha/gamma agonist for the treatment of type II diabetes | | Descriptor: | (2S)-2-methoxy-3-{4-[2-(5-methyl-2-phenyl-1,3-oxazol-4-yl)ethoxy]-1-benzothiophen-7-yl}propanoic acid, Nuclear receptor coactivator 1, Peroxisome proliferator-activated receptor alpha | | Authors: | Benz, J, Bernardeau, A, Binggeli, A, Blum, D, Boehringer, M, Grether, U, Hilpert, H, Kuhn, B, Maerki, H.P, Meyer, M, Puentener, K, Raab, S, Ruf, A, Schlatter, D, Gsell, B, Stihle, M, Mohr, P. | | Deposit date: | 2009-02-12 | | Release date: | 2009-06-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Aleglitazar, a new, potent, and balanced dual PPARalpha/gamma agonist for the treatment of type II diabetes.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
5QD1
 
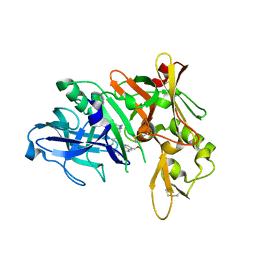 | | Crystal structure of BACE complex with BMC011 | | Descriptor: | (10S,12S)-12-[(1R)-1-hydroxy-2-({[3-(propan-2-yl)phenyl]methyl}amino)ethyl]-17-(methoxymethyl)-10-methyl-7-oxa-2,13-diazabicyclo[13.3.1]nonadeca-1(19),15,17-trien-14-one, Beta-secretase 1 | | Authors: | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-12-01 | | Release date: | 2020-06-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
4ZED
 
 | |
3JU3
 
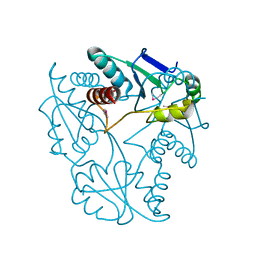 | | Crystal structure of alpha chain of probable 2-oxoacid ferredoxin oxidoreductase from Thermoplasma acidophilum | | Descriptor: | 1,2-ETHANEDIOL, Probable 2-oxoacid ferredoxin oxidoreductase, alpha chain | | Authors: | Chang, C, Marshall, N, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-09-14 | | Release date: | 2009-09-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of alpha chain of probable 2-oxoacid ferredoxin oxidoreductase from Thermoplasma acidophilum
To be Published
|
|
5QDM
 
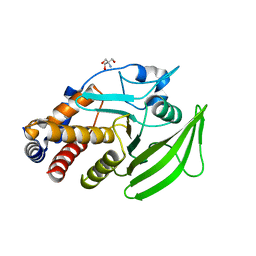 | | PanDDA analysis group deposition -- Crystal structure of PTP1B in complex with compound_FMOPL000074a | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[4-(1~{H}-pyrazol-3-yl)phenoxy]pyrimidine, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Keedy, D.A, Hill, Z.B, Biel, J.T, Kang, E, Rettenmaier, T.J, Brandao-Neto, J, von Delft, F, Wells, J.A, Fraser, J.S. | | Deposit date: | 2018-08-30 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.647 Å) | | Cite: | An expanded allosteric network in PTP1B by multitemperature crystallography, fragment screening, and covalent tethering.
Elife, 7, 2018
|
|
2VW8
 
 | | Crystal Structure of Quinolone signal response protein pqsE from Pseudomonas aeruginosa | | Descriptor: | 1,2-ETHANEDIOL, CACODYLATE ION, FE (II) ION, ... | | Authors: | Carter, L.G, Johnson, K.A, Liu, H, Mcmahon, S.A, Oke, M, Naismith, J.H, White, M.F. | | Deposit date: | 2008-06-17 | | Release date: | 2010-07-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genomics, 11, 2010
|
|
3K54
 
 | | Structures of human Bruton's tyrosine kinase in active and inactive conformations suggests a mechanism of activation for TEC family kinases. | | Descriptor: | N-(2-CHLORO-6-METHYLPHENYL)-2-({6-[4-(2-HYDROXYETHYL)PIPERAZIN-1-YL]-2-METHYLPYRIMIDIN-4-YL}AMINO)-1,3-THIAZOLE-5-CARBOXAMIDE, Tyrosine-protein kinase BTK | | Authors: | Marcotte, D.J, Liu, Y.-T, Arduini, R.M, Hession, C.A, Miatkowski, K, Wildes, C.P, Cullen, P.F, Hopkins, B.T, Mertsching, E, Jenkins, T.J, Romanowski, M.J, Baker, D.P, Silvian, L.F. | | Deposit date: | 2009-10-06 | | Release date: | 2010-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structures of human Bruton's tyrosine kinase in active and inactive conformations suggest a mechanism of activation for TEC family kinases.
Protein Sci., 19, 2010
|
|
