2C78
 
 | | EF-Tu complexed with a GTP analog and the antibiotic pulvomycin | | Descriptor: | (1S,2S,3E,5E,7E,10S,11S,12S)-12-[(2R,4E,6E,8Z,10R,12E,14E,16Z,18S,19Z)-10,18-DIHYDROXY-12,16,19-TRIMETHYL-11,22-DIOXOOX ACYCLODOCOSA-4,6,8,12,14,16,19-HEPTAEN-2-YL]-2,11-DIHYDROXY-1,10-DIMETHYL-9-OXOTRIDECA-3,5,7-TRIEN-1-YL 6-DEOXY-2,4-DI-O-METHYL-BETA-L-GALACTOPYRANOSIDE, ELONGATION FACTOR TU-A, MAGNESIUM ION, ... | | Authors: | Parmeggiani, A, Krab, I.M, Okamura, S, Nielsen, R.C, Nyborg, J, Nissen, P. | | Deposit date: | 2005-11-18 | | Release date: | 2006-03-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis of the action of pulvomycin and GE2270 A on elongation factor Tu.
Biochemistry, 45, 2006
|
|
2ECZ
 
 | | Solution structure of the SH3 domain of Sorbin and SH3 domain-containing protein 1 | | Descriptor: | Sorbin and SH3 domain-containing protein 1 | | Authors: | Abe, H, Miyamoto, K, Tochio, N, Saito, K, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-02-14 | | Release date: | 2008-02-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the SH3 domain of Sorbin and SH3 domain-containing protein 1
To be Published
|
|
6E21
 
 | | Joint X-ray/neutron structure of PKAc with products Sr2-ADP and phosphorylated peptide SP20 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, STRONTIUM ION, cAMP-dependent protein kinase catalytic subunit alpha, ... | | Authors: | Kovalevsky, A, Gerlits, O.O, Taylor, S. | | Deposit date: | 2018-07-10 | | Release date: | 2019-04-03 | | Last modified: | 2023-10-25 | | Method: | NEUTRON DIFFRACTION (2 Å), X-RAY DIFFRACTION | | Cite: | Zooming in on protons: Neutron structure of protein kinase A trapped in a product complex.
Sci Adv, 5, 2019
|
|
6EDE
 
 | | tRNA 2'-phosphotransferase | | Descriptor: | Probable RNA 2'-phosphotransferase, [[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{S},4~{R},5~{R})-3,4-bis(oxidanyl)-5-phosphonooxy-oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Shuman, S, Goldgur, Y, Banerjee, A. | | Deposit date: | 2018-08-09 | | Release date: | 2019-03-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.553 Å) | | Cite: | Structure of tRNA splicing enzyme Tpt1 illuminates the mechanism of RNA 2'-PO4recognition and ADP-ribosylation.
Nat Commun, 10, 2019
|
|
6E3A
 
 | | tRNA 2'-phosphotransferase | | Descriptor: | COENZYME A, Probable RNA 2'-phosphotransferase, [[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{S},4~{R},5~{R})-3,4-bis(oxidanyl)-5-phosphonooxy-oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Shuman, S, Goldgur, Y, Banerjee, A. | | Deposit date: | 2018-07-13 | | Release date: | 2019-03-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of tRNA splicing enzyme Tpt1 illuminates the mechanism of RNA 2'-PO4recognition and ADP-ribosylation.
Nat Commun, 10, 2019
|
|
2B2W
 
 | | Tandem chromodomains of human CHD1 complexed with Histone H3 Tail containing trimethyllysine 4 | | Descriptor: | Chromodomain-helicase-DNA-binding protein 1, Histone H3 | | Authors: | Flanagan IV, J.F, Mi, L.-Z, Chruszcz, M, Cymborowski, M, Clines, K.L, Kim, Y, Minor, W, Rastinejad, F, Khorasanizadeh, S. | | Deposit date: | 2005-09-19 | | Release date: | 2005-12-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Double chromodomains cooperate to recognize the methylated histone H3 tail.
Nature, 438, 2005
|
|
2B2Y
 
 | | Tandem chromodomains of human CHD1 | | Descriptor: | Chromodomain-helicase-DNA-binding protein 1 | | Authors: | Flanagan IV, J.F, Mi, L.-Z, Chruszcz, M, Cymborowski, M, Clines, K.L, Kim, Y, Minor, W, Rastinejad, F, Khorasanizadeh, S. | | Deposit date: | 2005-09-19 | | Release date: | 2005-12-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Double chromodomains cooperate to recognize the methylated histone H3 tail.
Nature, 438, 2005
|
|
2B2V
 
 | | Crystal structure analysis of human CHD1 chromodomains 1 and 2 bound to histone H3 resi 1-15 MeK4 | | Descriptor: | Chromodomain-helicase-DNA-binding protein 1, Histone H3 | | Authors: | Flanagan IV, J.F, Mi, L.-Z, Chruszcz, M, Cymborowski, M, Clines, K.L, Kim, Y, Minor, W, Rastinejad, F, Khorasanizadeh, S. | | Deposit date: | 2005-09-19 | | Release date: | 2005-12-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Double chromodomains cooperate to recognize the methylated histone H3 tail.
Nature, 438, 2005
|
|
2B2U
 
 | | Tandem chromodomains of human CHD1 complexed with Histone H3 Tail containing trimethyllysine 4 and dimethylarginine 2 | | Descriptor: | Chromodomain-helicase-DNA-binding protein 1, Histone H3 | | Authors: | Flanagan IV, J.F, Mi, L.-Z, Chruszcz, M, Cymborowski, M, Clines, K.L, Kim, Y, Minor, W, Rastinejad, F, Khorasanizadeh, S. | | Deposit date: | 2005-09-19 | | Release date: | 2005-12-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Double chromodomains cooperate to recognize the methylated histone H3 tail.
Nature, 438, 2005
|
|
2FAQ
 
 | | Crystal Structure of Pseudomonas aeruginosa LigD polymerase domain with ATP and Manganese | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, SULFATE ION, ... | | Authors: | Zhu, H, Nandakumar, J, Aniukwu, J, Wang, L.K, Glickman, M.S, Lima, C.D, Shuman, S. | | Deposit date: | 2005-12-07 | | Release date: | 2006-05-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Atomic structure and nonhomologous end-joining function of the polymerase component of bacterial DNA ligase D
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
2DGJ
 
 | | Crystal structure of EbhA (756-1003 domain) from Staphylococcus aureus | | Descriptor: | ACETIC ACID, GLYCEROL, SULFATE ION, ... | | Authors: | Tanaka, Y, Yao, M, Kuroda, M, Watanabe, N, Ohta, T, Tanaka, I. | | Deposit date: | 2006-03-14 | | Release date: | 2007-03-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A helical string of alternately connected three-helix bundles for the cell wall-associated adhesion protein Ebh from Staphylococcus aureus
Structure, 16, 2008
|
|
2FAO
 
 | | Crystal Structure of Pseudomonas aeruginosa LigD polymerase domain | | Descriptor: | SULFATE ION, probable ATP-dependent DNA ligase | | Authors: | Zhu, H, Nandakumar, J, Aniukwu, J, Wang, L.K, Glickman, M.S, Lima, C.D, Shuman, S. | | Deposit date: | 2005-12-07 | | Release date: | 2006-05-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Atomic structure and nonhomologous end-joining function of the polymerase component of bacterial DNA ligase D
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
2ERZ
 
 | | Crystal Structure of c-AMP Dependent Kinase (PKA) bound to hydroxyfasudil | | Descriptor: | 1-(1-HYDROXY-5-ISOQUINOLINESULFONYL)HOMOPIPERAZINE, cAMP-dependent protein kinase inhibitor, alpha form, ... | | Authors: | Jacobs, M. | | Deposit date: | 2005-10-25 | | Release date: | 2005-11-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Structure of Dimeric ROCK I Reveals the Mechanism for Ligand Selectivity.
J.Biol.Chem., 281, 2006
|
|
4V89
 
 | | Crystal Structure of Release Factor RF3 Trapped in the GTP State on a Rotated Conformation of the Ribosome (without viomycin) | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Zhou, J, Lancaster, L, Trakhanov, S, Noller, H.F. | | Deposit date: | 2011-11-17 | | Release date: | 2014-07-09 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Crystal structure of release factor RF3 trapped in the GTP state on a rotated conformation of the ribosome.
Rna, 18, 2012
|
|
4V8G
 
 | | Crystal structure of RMF bound to the 70S ribosome. | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S Ribosomal Protein S10, ... | | Authors: | Polikanov, Y.S, Blaha, G.M, Steitz, T.A. | | Deposit date: | 2011-12-11 | | Release date: | 2014-07-09 | | Last modified: | 2014-12-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | How hibernation factors RMF, HPF, and YfiA turn off protein synthesis.
Science, 336, 2012
|
|
8AYO
 
 | | Open state GluA1/A2 AMPA receptor in complex with TARP gamma 8 and ligand JNJ-61432059 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 5-[2-(4-fluorophenyl)-7-(4-oxidanylpiperidin-1-yl)pyrazolo[1,5-c]pyrimidin-3-yl]-1,3-dihydroindol-2-one, ... | | Authors: | Zhang, D, Lape, R, Shaikh, S, Kohegyi, B, Watson, J.F, Cais, O, Nakagawa, T, Greger, I.H. | | Deposit date: | 2022-09-02 | | Release date: | 2023-04-19 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Modulatory mechanisms of TARP gamma 8-selective AMPA receptor therapeutics.
Nat Commun, 14, 2023
|
|
4V3V
 
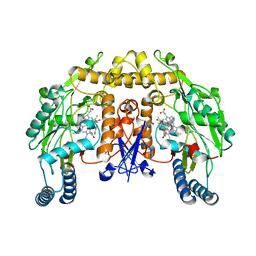 | | Structure of rat neuronal nitric oxide synthase heme domain in complex with N-(2-(1H-imidazol-1-yl)-4-pyrimidylmethyl)-3-(3- fluorophenyl)propan-1-amine | | Descriptor: | 3-(3-fluorophenyl)-N-{[2-(1H-imidazol-1-yl)pyrimidin-4-yl]methyl}propan-1-amine, 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2014-10-20 | | Release date: | 2014-12-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Novel 2,4-Disubstituted Pyrimidines as Potent, Selective, and Cell-Permeable Inhibitors of Neuronal Nitric Oxide Synthase.
J.Med.Chem., 58, 2015
|
|
6IDF
 
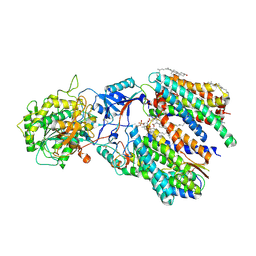 | | Cryo-EM structure of gamma secretase in complex with a Notch fragment | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yang, G, Zhou, R, Zhou, Q, Guo, X, Yan, C, Ke, M, Lei, J, Shi, Y. | | Deposit date: | 2018-09-09 | | Release date: | 2018-12-26 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis of Notch recognition by human gamma-secretase
Nature, 565, 2019
|
|
4W9Y
 
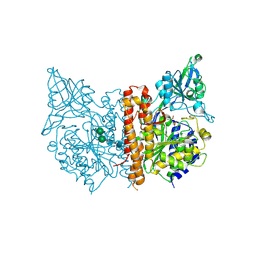 | |
6IID
 
 | | Human EXOG-H140A in complex with RNA-DNA chimeric duplex | | Descriptor: | DNA (5'-D(*CP*GP*TP*GP*AP*CP*AP*TP*CP*CP*CP*G)-3'), DNA/RNA (5'-R(P*CP*GP*GP*GP*A)-D(P*T)-R(P*G)-D(P*T)-R(P*CP*AP*CP*G)-3'), MAGNESIUM ION, ... | | Authors: | Wu, C.C, Lin, J.L.J, Yuan, H.S. | | Deposit date: | 2018-10-04 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.986 Å) | | Cite: | A unique exonuclease ExoG cleaves between RNA and DNA in mitochondrial DNA replication.
Nucleic Acids Res., 47, 2019
|
|
4NE5
 
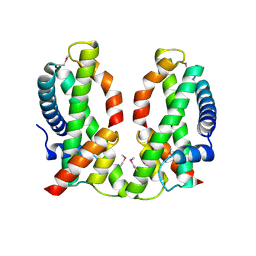 | | Human MHF1-MHF2 complex | | Descriptor: | Centromere protein S, Centromere protein X | | Authors: | Zhao, Q, Saro, D, Sachpatzidis, A, Sung, P, Xiong, Y. | | Deposit date: | 2013-10-28 | | Release date: | 2013-12-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The MHF complex senses branched DNA by binding a pair of crossover DNA duplexes.
Nat Commun, 5, 2014
|
|
4V54
 
 | | Crystal structure of the bacterial ribosome from Escherichia coli in complex with ribosome recycling factor (RRF). | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Borovinskaya, M.A, Pai, R.D, Zhang, W, Schuwirth, B.-S, Holton, J.M, Hirokawa, G, Kaji, H, Kaji, A, Cate, J.H.D. | | Deposit date: | 2007-06-16 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis for aminoglycoside inhibition of bacterial ribosome recycling.
Nat.Struct.Mol.Biol., 14, 2007
|
|
4V6R
 
 | | Structural characterization of mRNA-tRNA translocation intermediates (class 6 of the six classes) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Agirrezabala, X, Liao, H, Schreiner, E, Fu, J, Ortiz-Meoz, R.F, Schulten, K, Green, R, Frank, J. | | Deposit date: | 2011-12-08 | | Release date: | 2014-07-09 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (11.5 Å) | | Cite: | Structural characterization of mRNA-tRNA translocation intermediates.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4V76
 
 | | E. coli 70S-fMetVal-tRNAVal-tRNAfMet complex in intermediate post-translocation state (post2a) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Blau, C, Bock, L.V, Schroder, G.F, Davydov, I, Fischer, N, Stark, H, Rodnina, M.V, Vaiana, A.C, Grubmuller, H. | | Deposit date: | 2013-10-14 | | Release date: | 2014-07-09 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (17 Å) | | Cite: | Energy barriers and driving forces in tRNA translocation through the ribosome.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4V4Q
 
 | | Crystal structure of the bacterial ribosome from Escherichia coli at 3.5 A resolution. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Schuwirth, B.S, Borovinskaya, M.A, Hau, C.W, Zhang, W, Vila-Sanjurjo, A, Holton, J.M, Cate, J.H.D. | | Deposit date: | 2005-08-30 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.46 Å) | | Cite: | Structures of the bacterial ribosome at 3.5 A resolution.
Science, 310, 2005
|
|
