6JW7
 
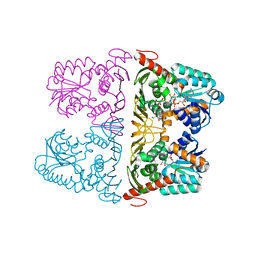 | | The crystal structure of KanD2 in complex with NADH and 3"-deamino-3"-hydroxykanamycin A | | Descriptor: | (2R,3S,4S,5R,6R)-2-(aminomethyl)-6-[(1R,2S,3S,4R,6S)-4,6-bis(azanyl)-3-[(2S,3R,4S,5S,6R)-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-2-oxidanyl-cyclohexyl]oxy-oxane-3,4,5-triol, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Dehydrogenase | | Authors: | Kudo, F, Kitayama, Y, Miyanaga, A, Hirayama, A, Eguchi, T. | | Deposit date: | 2019-04-18 | | Release date: | 2020-04-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Biochemical and structural analysis of a dehydrogenase, KanD2, and an aminotransferase, KanS2, that are responsible for the construction of the kanosamine moiety in kanamycin biosynthesis.
Biochemistry, 59, 2020
|
|
6FZQ
 
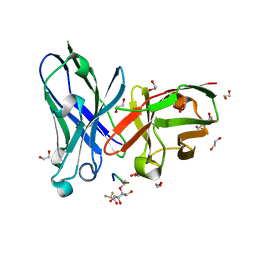 | | Crystal structure of scFv-SM3 in complex with compound 3 | | Descriptor: | 1,2-ETHANEDIOL, 2-deoxy-2-[(difluoroacetyl)amino]-alpha-D-galactopyranose, Mucin-1, ... | | Authors: | Bermejo, I.A, Usabiaga, I, Companon, I, Castro-Lopez, J, Insausti, A, Fernandez, J.A, Avenoza, A, Busto, J.H, Jimenez-Barbero, J, Asensio, J.L, Jimenez-Oses, G, Peregrina, J.M, Hurtado-Guerrero, R, Cocinero, E.J, Corzana, F. | | Deposit date: | 2018-03-15 | | Release date: | 2019-02-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Water Sculpts the Distinctive Shapes and Dynamics of the Tumor-Associated Carbohydrate Tn Antigens: Implications for Their Molecular Recognition.
J.Am.Chem.Soc., 140, 2018
|
|
6VH1
 
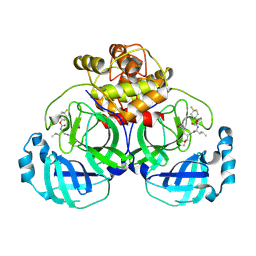 | | 2.30 A resolution structure of MERS 3CL protease in complex with inhibitor 6h | | Descriptor: | N~2~-{[(4,4-difluorocyclohexyl)oxy]carbonyl}-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide, Orf1a protein | | Authors: | Lovell, S, Battaile, K.P, Kashipathy, M.M, Rathnayake, A.D, Zheng, J, Kim, Y, Nguyen, H.N, Chang, K.O, Groutas, W.C. | | Deposit date: | 2020-01-09 | | Release date: | 2020-08-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | 3C-like protease inhibitors block coronavirus replication in vitro and improve survival in MERS-CoV-infected mice.
Sci Transl Med, 12, 2020
|
|
3ILA
 
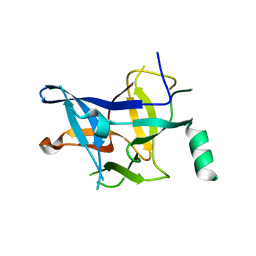 | |
4CMF
 
 | | The (R)-selective transaminase from Nectria haematococca with inhibitor bound | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-[O-PHOSPHONOPYRIDOXYL]--AMINO-BENZOIC ACID, AMINOTRANSFERASE, ... | | Authors: | Sayer, C, Isupov, M, Martinez-Torres, R.J, Richter, N, Hailes, H.C, Ward, J, Littlechild, J. | | Deposit date: | 2014-01-16 | | Release date: | 2014-03-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Substrate Specificity, Enantioselectivity and Structure of the (R)-Selective Amine:Pyruvate Transaminase from Nectria Haematococca.
FEBS J., 281, 2014
|
|
4CSJ
 
 | | The discovery of potent selective glucocorticoid receptor modulators, suitable for inhalation | | Descriptor: | 1,2-ETHANEDIOL, GLUCOCORTICOID RECEPTOR, N-[(2S)-1-[[1-(4-fluorophenyl)indazol-4-yl]amino]propan-2-yl]-2,4,6-trimethyl-benzenesulfonamide, ... | | Authors: | Edman, K, Ahlgren, R, Bengtsson, M, Bladh, H, Backstrom, S, Dahmen, J, Henriksson, K, Hillertz, P, Hulikal, V, Jerre, A, Kinchin, L, Kase, C, Lepisto, M, Mile, I, Nilsson, S, Smailagic, A, Taylor, J, Tjornebo, A, Wissler, L, Hansson, T. | | Deposit date: | 2014-03-07 | | Release date: | 2014-05-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Discovery of Potent and Selective Non-Steroidal Glucocorticoid Receptor Modulators, Suitable for Inhalation.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
5TYE
 
 | | DNA Polymerase Mu Product Complex, 10 mM Mg2+ (60 min) | | Descriptor: | 1,2-ETHANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Jamsen, J.A, Wilson, S.H. | | Deposit date: | 2016-11-19 | | Release date: | 2017-08-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.047 Å) | | Cite: | Time-lapse crystallography snapshots of a double-strand break repair polymerase in action.
Nat Commun, 8, 2017
|
|
6JTA
 
 | | Crystal Structure of D464A L465A mutant of FGAM Synthetase | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, GLUTAMINE, ... | | Authors: | Sharma, N, Ahalawat, N, Sandhu, P, Mondal, J, Anand, R. | | Deposit date: | 2019-04-10 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Role of allosteric switches and adaptor domains in long-distance cross-talk and transient tunnel formation.
Sci Adv, 6, 2020
|
|
7GD5
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with ALP-POS-d2866bdf-1 (Mpro-x10876) | | Descriptor: | 2-(1H-benzotriazol-1-yl)-N-[4-(dimethylamino)phenyl]-N-[(thiophen-3-yl)methyl]acetamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.809 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
4PTF
 
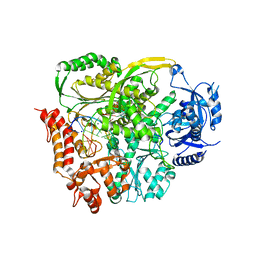 | | Ternary crystal structure of yeast DNA polymerase epsilon with template G | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, 5'-D(*AP*TP*CP*CP*TP*CP*CP*CP*CP*TP*AP*(DOC))-3', ... | | Authors: | Jain, R, Rajashankar, K.R, Buku, A, Johnson, R.E, Prakash, L, Prakash, S, Aggarwal, A.K. | | Deposit date: | 2014-03-10 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.809 Å) | | Cite: | Crystal Structure of Yeast DNA Polymerase epsilon Catalytic Domain.
Plos One, 9, 2014
|
|
3IK1
 
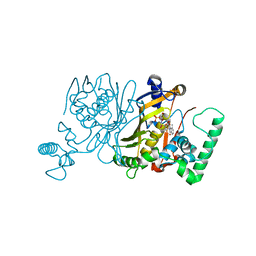 | | Lactobacillus casei Thymidylate Synthase in Ternary Complex with dUMP and the Phtalimidic Derivative 20C | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 2-hydroxy-4-(4-methyl-1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)benzoic acid, Thymidylate synthase | | Authors: | Pozzi, C, Cancian, L, Leone, R, Luciani, R, Ferrari, S, Mangani, S, Costi, M.P. | | Deposit date: | 2009-08-05 | | Release date: | 2010-08-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Identification of the binding modes of N-phenylphthalimides inhibiting bacterial thymidylate synthase through X-ray crystallography screening
J.Med.Chem., 54, 2011
|
|
5Y3N
 
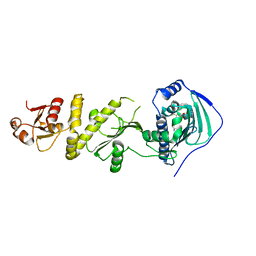 | | Structure of TRAP1 complexed with DN401 | | Descriptor: | 1-[(6-bromanyl-1,3-benzodioxol-5-yl)methyl]-4-chloranyl-pyrazolo[3,4-d]pyrimidin-6-amine, Heat shock protein 75 kDa, mitochondrial | | Authors: | Jeong, H, Park, H.K, Kang, S, Kang, B.H, Lee, C. | | Deposit date: | 2017-07-29 | | Release date: | 2017-08-30 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Paralog Specificity Determines Subcellular Distribution, Action Mechanism, and Anticancer Activity of TRAP1 Inhibitors.
J. Med. Chem., 60, 2017
|
|
7GGN
 
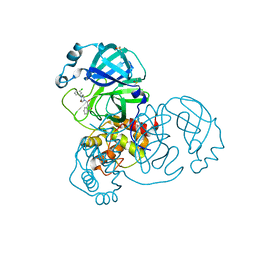 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with ERI-UCB-d6de1f3c-1 (Mpro-x12679) | | Descriptor: | 3C-like proteinase, 4-[4-(3-chlorophenyl)-3-oxopiperazine-1-carbonyl]quinolin-2(1H)-one, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.928 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
2YJC
 
 | | CATHEPSIN L WITH A NITRILE INHIBITOR | | Descriptor: | (2S,4R)-1-[1-(4-chlorophenyl)cyclopropyl]carbonyl-4-(2-chlorophenyl)sulfonyl-N-[1-(iminomethyl)cyclopropyl]pyrrolidine-2-carboxamide, CATHEPSIN L1 | | Authors: | Banner, D.W, Benz, J.M, Haap, W. | | Deposit date: | 2011-05-19 | | Release date: | 2011-11-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Halogen Bonding at the Active Sites of Human Cathepsin L and Mek1 Kinase: Efficient Interactions in Different Environments.
Chemmedchem, 6, 2011
|
|
4PVG
 
 | |
4D2J
 
 | | Crystal structure of F16BP Aldolase from Toxoplasma gondii (TgALD1) | | Descriptor: | ACETATE ION, BETA-MERCAPTOETHANOL, FRUCTOSE-BISPHOSPHATE ALDOLASE, ... | | Authors: | Tonkin, M.L, Ramaswamy, R, Halavaty, A.S, Ruan, J, Igarashi, M, Ngo, H.M, Boulanger, M.J. | | Deposit date: | 2014-05-09 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and Functional Divergence of the Aldolase Fold in Toxoplasma Gondii.
J.Mol.Biol., 427, 2015
|
|
5A3C
 
 | | Crystal structure of the ADP-ribosylating sirtuin (SirTM) from Streptococcus pyogenes in complex with NAD | | Descriptor: | 1,2-ETHANEDIOL, GLYCINE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Rack, J.G.M, Morra, R, Barkauskaite, E, Kraehenbuehl, R, Ariza, A, Qu, Y, Ortmayer, M, Leidecker, O, Cameron, D.R, Matic, I, Peleg, A.Y, Leys, D, Traven, A, Ahel, I. | | Deposit date: | 2015-05-28 | | Release date: | 2015-07-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Identification of a Class of Protein Adp-Ribosylating Sirtuins in Microbial Pathogens.
Mol.Cell, 59, 2015
|
|
6G38
 
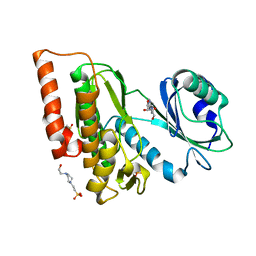 | | Crystal structure of haspin in complex with tubercidin | | Descriptor: | '2-(4-AMINO-PYRROLO[2,3-D]PYRIMIDIN-7-YL)-5-HYDROXYMETHYL-TETRAHYDRO-FURAN-3,4-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DIMETHYL SULFOXIDE, ... | | Authors: | Heroven, C, Chaikuad, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-03-24 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Halogen-Aromatic pi Interactions Modulate Inhibitor Residence Times.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
6G3C
 
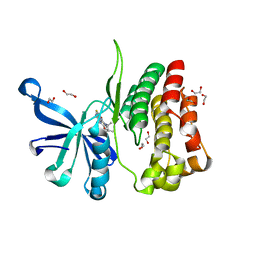 | | Crystal Structure of JAK2-V617F pseudokinase domain in complex with Compound 2 | | Descriptor: | 1,2-ETHANEDIOL, 2-[[3,5-bis(fluoranyl)-4-oxidanyl-phenyl]amino]-5,7,7-trimethyl-8-(3-methylbutyl)pteridin-6-one, Tyrosine-protein kinase | | Authors: | Dekker, C, Hinniger, A. | | Deposit date: | 2018-03-24 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery and Structural Characterization of ATP-Site Ligands for the Wild-Type and V617F Mutant JAK2 Pseudokinase Domain.
Acs Chem.Biol., 14, 2019
|
|
5BW5
 
 | |
6K5J
 
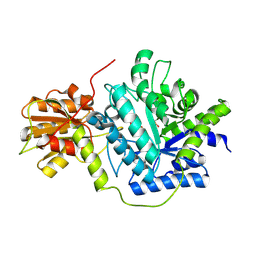 | | Structure of a glycoside hydrolase family 3 beta-N-acetylglucosaminidase from Paenibacillus sp. str. FPU-7 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GH3 beta-N-acetylglucosaminidase, GLYCEROL | | Authors: | Itoh, T, Araki, T, Nishiyama, T, Hibi, T, Kimoto, H. | | Deposit date: | 2019-05-29 | | Release date: | 2019-09-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.903 Å) | | Cite: | Structural and functional characterization of a glycoside hydrolase family 3 beta-N-acetylglucosaminidase from Paenibacillus sp. str. FPU-7.
J.Biochem., 166, 2019
|
|
4ZTQ
 
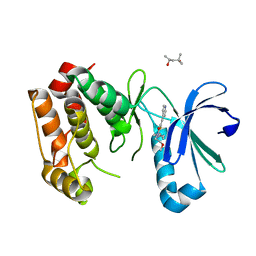 | | Human Aurora A catalytic domain bound to FK932 | | Descriptor: | (2Z,5Z)-2-[(4-ethylphenyl)imino]-3-(2-methoxyethyl)-5-(pyridin-4-ylmethylidene)-1,3-thiazolidin-4-one, (4S)-2-METHYL-2,4-PENTANEDIOL, Aurora kinase A | | Authors: | Marcaida, M.J, Kilchmann, F, Schick, T, Reymond, J.L. | | Deposit date: | 2015-05-14 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of a Selective Aurora A Kinase Inhibitor by Virtual Screening.
J.Med.Chem., 59, 2016
|
|
5BY5
 
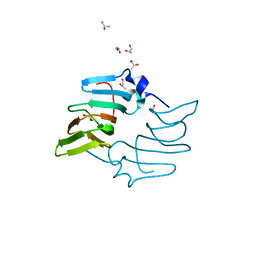 | | High resolution structure of the ectoine synthase from the cold-adapted marine bacterium Sphingopyxis alaskensis | | Descriptor: | L-ectoine synthase, S-1,2-PROPANEDIOL | | Authors: | Widderich, N, Kobus, S, Hoeppner, A, Bremer, E, Smits, S.H.J. | | Deposit date: | 2015-06-10 | | Release date: | 2016-04-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Biochemistry and Crystal Structure of Ectoine Synthase: A Metal-Containing Member of the Cupin Superfamily.
Plos One, 11, 2016
|
|
3JS5
 
 | |
5Y2Y
 
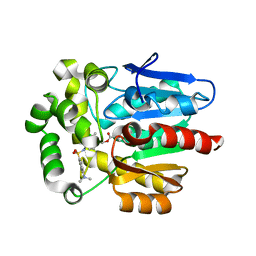 | | Crystal structure of HaloTag (M175C) complexed with dansyl-PEG2-HaloTag ligand | | Descriptor: | 5-(dimethylamino)-~{N}-[2-(2-hexoxyethoxy)ethyl]naphthalene-1-sulfonamide, CHLORIDE ION, Haloalkane dehalogenase | | Authors: | Lee, H, Kang, M, Rhee, H, Lee, C. | | Deposit date: | 2017-07-27 | | Release date: | 2017-09-06 | | Last modified: | 2025-09-17 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structure-guided synthesis of a protein-based fluorescent sensor for alkyl halides
Chem. Commun. (Camb.), 53, 2017
|
|
