7BAR
 
 | |
7SVY
 
 | | MicroED structure of proteinase K from a 130 nm thick lamella measured at 120 kV | | Descriptor: | Proteinase K | | Authors: | Martynowycz, M.W, Clabbers, M.T.B, Unge, J, Hattne, J, Gonen, T. | | Deposit date: | 2021-11-19 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-18 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.3 Å) | | Cite: | Benchmarking the ideal sample thickness in cryo-EM.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6J43
 
 | | Proteinase K determined by PAL-XFEL | | Descriptor: | CALCIUM ION, Proteinase K | | Authors: | Lee, S.J, Park, J. | | Deposit date: | 2019-01-07 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Application of a high-throughput microcrystal delivery system to serial femtosecond crystallography.
J.Appl.Crystallogr., 53, 2020
|
|
8F8R
 
 | | Cryo-EM structure of the free barbed end of F-actin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Carman, P.J, Barrie, K.R, Dominguez, R. | | Deposit date: | 2022-11-22 | | Release date: | 2023-06-07 | | Last modified: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structures of the free and capped ends of the actin filament.
Science, 380, 2023
|
|
7SWB
 
 | | MicroED structure of proteinase K from a 360 nm thick lamella measured at 300 kV | | Descriptor: | Proteinase K | | Authors: | Martynowycz, M.W, Clabbers, M.T.B, Unge, J, Hattne, J, Gonen, T. | | Deposit date: | 2021-11-19 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-18 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.05 Å) | | Cite: | Benchmarking the ideal sample thickness in cryo-EM.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
4NGY
 
 | | Dialyzed HEW lysozyme batch crystallized in 0.75 M YbCl3 and collected at 100 K | | Descriptor: | CHLORIDE ION, Lysozyme C, YTTERBIUM (III) ION | | Authors: | Benas, P, Legrand, L, Ries-Kautt, M. | | Deposit date: | 2013-11-03 | | Release date: | 2014-05-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Weak protein-cationic co-ion interactions addressed by X-ray crystallography and mass spectrometry.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
7SW2
 
 | | MicroED structure of proteinase K from a 130 nm thick lamella measured at 200 kV | | Descriptor: | Proteinase K | | Authors: | Martynowycz, M.W, Clabbers, M.T.B, Unge, J, Hattne, J, Gonen, T. | | Deposit date: | 2021-11-19 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-18 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.95 Å) | | Cite: | Benchmarking the ideal sample thickness in cryo-EM.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
8F8P
 
 | | Cryo-EM structure of F-actin in the ADP state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Carman, P.J, Barrie, K.R, Dominguez, R. | | Deposit date: | 2022-11-22 | | Release date: | 2023-06-07 | | Last modified: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (2.26 Å) | | Cite: | Structures of the free and capped ends of the actin filament.
Science, 380, 2023
|
|
7BB0
 
 | | Crystal structure of the cAMP-dependent protein kinase A cocrystallized with NAT22-366511 and PKI (5-24) | | Descriptor: | (R)-3,3-dimethyl-5-oxo-5-(6-(3-(pyridin-4-yl)-1,2,4-oxadiazol-5-yl)-1,4,6,7-tetrahydro-5H-imidazo[4,5-c]pyridin-5-yl)pentanoic acid, cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase inhibitor alpha | | Authors: | Oebbeke, M, Mueller, J, Glinca, S, Heine, A, Klebe, G. | | Deposit date: | 2020-12-16 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of the cAMP-dependent protein kinase A cocrystallized with NAT22-366511 and PKI (5-24)
To Be Published
|
|
7SW8
 
 | | MicroED structure of proteinase K from a 150 nm thick lamella measured at 300 kV | | Descriptor: | Proteinase K | | Authors: | Martynowycz, M.W, Clabbers, M.T.B, Unge, J, Hattne, J, Gonen, T. | | Deposit date: | 2021-11-19 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-18 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.9 Å) | | Cite: | Benchmarking the ideal sample thickness in cryo-EM.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
8F8S
 
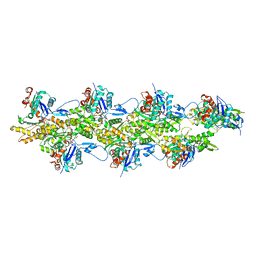 | | Cryo-EM structure of the free pointed end of F-actin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Carman, P.J, Barrie, K.R, Dominguez, R. | | Deposit date: | 2022-11-22 | | Release date: | 2023-06-07 | | Last modified: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Structures of the free and capped ends of the actin filament.
Science, 380, 2023
|
|
6JFV
 
 | | The crystal structure of 2B-2B complex from keratins 5 and 14 (C367A mutant of K14) | | Descriptor: | Keratin, type I cytoskeletal 14, type II cytoskeletal 5 | | Authors: | Kim, M.S, Lee, C.H, Coulombe, P.A, Leahy, D.J. | | Deposit date: | 2019-02-12 | | Release date: | 2020-01-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-Function Analyses of a Keratin Heterotypic Complex Identify Specific Keratin Regions Involved in Intermediate Filament Assembly.
Structure, 28, 2020
|
|
4NJ3
 
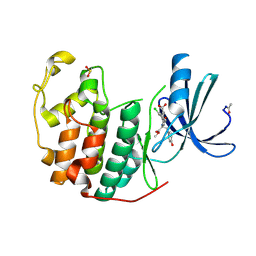 | |
7T1U
 
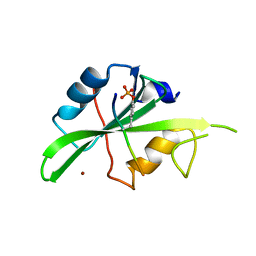 | | Crystal structure of a superbinder Src SH2 domain (sSrcF) in complex with a high affinity phosphopeptide | | Descriptor: | Proto-oncogene tyrosine-protein kinase Src, Synthetic phosphopeptide, ZINC ION | | Authors: | Martyn, G.D, Singer, A.U, Manczyk, N, Veggiani, G, Kurinov, I, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2021-12-02 | | Release date: | 2022-08-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Engineered SH2 Domains for Targeted Phosphoproteomics.
Acs Chem.Biol., 17, 2022
|
|
4NU1
 
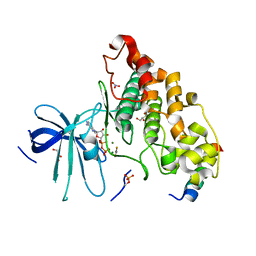 | | Crystal structure of a transition state mimic of the GSK-3/Axin complex bound to phosphorylated N-terminal auto-inhibitory pS9 peptide | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, Axin-1, ... | | Authors: | Chu, M.L.-H, Stamos, J.L, Enos, M.D, Shah, N, Weis, W.I. | | Deposit date: | 2013-12-03 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of GSK-3 inhibition by N-terminal phosphorylation and by the Wnt receptor LRP6.
Elife, 3, 2014
|
|
5UHM
 
 | |
6D0D
 
 | | X-ray crystal structure of wild type HIV-1 protease in complex with GRL-087-13 | | Descriptor: | (3aS,4S,7aR)-hexahydro-4H-furo[2,3-b]pyran-4-yl [(2S,3R)-1-(4-fluorophenyl)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}butan-2-yl]carbamate, Protease | | Authors: | Yedidi, R.S, Hayashi, H, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2018-04-10 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Novel Central Nervous System (CNS)-Targeting Protease Inhibitors for Drug-Resistant HIV Infection and HIV-Associated CNS Complications.
Antimicrob.Agents Chemother., 63, 2019
|
|
4NLJ
 
 | | Crystal structure of sheep beta-lactoglobulin (space group P1) | | Descriptor: | Beta-lactoglobulin-1/B, SULFATE ION | | Authors: | Loch, J.I, Molenda, M, Kopec, M, Swiatek, S, Lewinski, K. | | Deposit date: | 2013-11-14 | | Release date: | 2014-03-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of two crystal forms of sheep beta-lactoglobulin with EF-loop in closed conformation
Biopolymers, 101, 2014
|
|
5YIE
 
 | | Crystal Structure of KNI-10742 bound Plasmepsin II (PMII) from Plasmodium falciparum | | Descriptor: | (4R)-3-[(2S,3S)-3-[2-[4-[2-azanylethyl(ethyl)amino]-2,6-dimethyl-phenoxy]ethanoylamino]-2-oxidanyl-4-phenyl-butanoyl]-5,5-dimethyl-N-[(1S,2R)-2-oxidanyl-2,3-dihydro-1H-inden-1-yl]-1,3-thiazolidine-4-carboxamide, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, Plasmepsin II, ... | | Authors: | Mishra, V, Rathore, I, Bhaumik, P. | | Deposit date: | 2017-10-04 | | Release date: | 2018-07-11 | | Last modified: | 2019-05-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Deciphering the mechanism of potent peptidomimetic inhibitors targeting plasmepsins - biochemical and structural insights.
Febs J., 285, 2018
|
|
6CJ7
 
 | | Crystal structure of Manduca sexta Serine protease inhibitor (Serpin)-12 | | Descriptor: | Serpin-12 | | Authors: | Gulati, M, Hu, Y, Peng, S, Pathak, P.K, Wang, Y, Deng, J, Jiang, H. | | Deposit date: | 2018-02-26 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Manduca sexta serpin-12 controls the prophenoloxidase activation system in larval hemolymph.
Insect Biochem. Mol. Biol., 99, 2018
|
|
5YTU
 
 | |
7SW5
 
 | | MicroED structure of proteinase K from a 460 nm thick lamella measured at 200 kV | | Descriptor: | Proteinase K | | Authors: | Martynowycz, M.W, Clabbers, M.T.B, Unge, J, Hattne, J, Gonen, T. | | Deposit date: | 2021-11-19 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-18 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.95 Å) | | Cite: | Benchmarking the ideal sample thickness in cryo-EM.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6CJE
 
 | | Crystal Structure of Mnk2-D228G in complex with Inhibitor | | Descriptor: | 4-[(9H-purin-6-yl)amino]benzamide, MAP kinase-interacting serine/threonine-protein kinase 2, ZINC ION | | Authors: | Han, Q. | | Deposit date: | 2018-02-26 | | Release date: | 2018-05-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.36 Å) | | Cite: | Structure-based Design of Pyridone-Aminal eFT508 Targeting Dysregulated Translation by Selective Mitogen-activated Protein Kinase Interacting Kinases 1 and 2 (MNK1/2) Inhibition.
J. Med. Chem., 61, 2018
|
|
7SVZ
 
 | | MicroED structure of proteinase K from a 200 nm thick lamella measured at 120 kV | | Descriptor: | Proteinase K | | Authors: | Martynowycz, M.W, Clabbers, M.T.B, Unge, J, Hattne, J, Gonen, T. | | Deposit date: | 2021-11-19 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-18 | | Method: | ELECTRON CRYSTALLOGRAPHY (2 Å) | | Cite: | Benchmarking the ideal sample thickness in cryo-EM.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7SW6
 
 | | MicroED structure of proteinase K from a 260 nm thick lamella measured at 200 kV | | Descriptor: | Proteinase K | | Authors: | Martynowycz, M.W, Clabbers, M.T.B, Unge, J, Hattne, J, Gonen, T. | | Deposit date: | 2021-11-19 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-18 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.95 Å) | | Cite: | Benchmarking the ideal sample thickness in cryo-EM.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
