8T5M
 
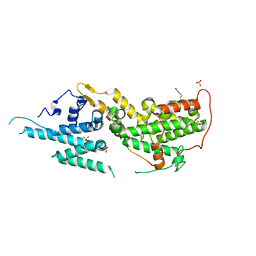 | | SOS2 crystal structure with fragment bound (compound 14) | | Descriptor: | 1,2-ETHANEDIOL, 4-[(1R,2S)-1-hydroxy-2-{[2-(4-hydroxyphenyl)ethyl]amino}propyl]phenol, SULFATE ION, ... | | Authors: | Gunn, R.J, Lawson, J.D, Ivetac, A, Ulaganathan, T, Coulombe, R, Fethiere, J. | | Deposit date: | 2023-06-14 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Discovery of Five SOS2 Fragment Hits with Binding Modes Determined by SOS2 X-Ray Cocrystallography.
J.Med.Chem., 67, 2024
|
|
8UFY
 
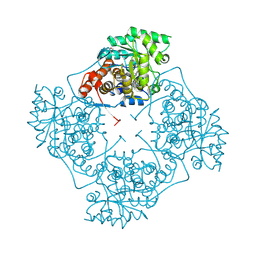 | | High Resolution structure of A. viridans Lactate Oxidase | | Descriptor: | 1,2-ETHANEDIOL, FLAVIN MONONUCLEOTIDE, L-lactate oxidase, ... | | Authors: | Lodowski, D.T. | | Deposit date: | 2023-10-05 | | Release date: | 2024-08-14 | | Last modified: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | An engineered lactate oxidase based electrochemical sensor for continuous detection of biomarker lactic acid in human sweat and serum.
Heliyon, 10, 2024
|
|
5A3X
 
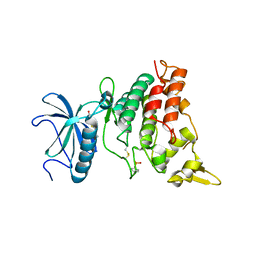 | | DYRK1A in complex with hydroxy benzothiazole fragment | | Descriptor: | DUAL SPECIFICITY TYROSINE-PHOSPHORYLATION-REGULATED KINASE 1A, N-(5-oxidanyl-1,3-benzothiazol-2-yl)ethanamide | | Authors: | Rothweiler, U. | | Deposit date: | 2015-06-03 | | Release date: | 2016-06-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Probing the ATP-Binding Pocket of Protein Kinase Dyrk1A with Benzothiazole Fragment Molecules
J.Med.Chem., 59, 2016
|
|
6YB8
 
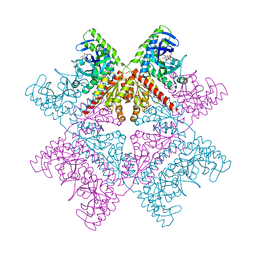 | | Human octameric PAICS in complex with CAIR and SAICAR | | Descriptor: | (2~{S})-2-[[5-azanyl-1-[(2~{R},3~{R},4~{S},5~{R})-3,4-bis(oxidanyl)-5-(phosphonooxymethyl)oxolan-2-yl]imidazol-4-yl]car bonylamino]butanedioic acid, 1,2-ETHANEDIOL, 5-AMINO-1-(5-O-PHOSPHONO-BETA-D-RIBOFURANOSYL)-1H-IMIDAZOLE-4-CARBOXYLIC ACID, ... | | Authors: | Skerlova, J, Unterlass, J, Gottmann, M, Homan, E, Helleday, T, Jemth, A.S, Stenmark, P. | | Deposit date: | 2020-03-16 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Crystal structures of human PAICS reveal substrate and product binding of an emerging cancer target.
J.Biol.Chem., 295, 2020
|
|
7Q0Y
 
 | | Crystal structure of CTX-M-14 in complex with Bortezomib | | Descriptor: | ACETATE ION, Beta-lactamase, CHLORIDE ION, ... | | Authors: | Werner, N, Perbandt, M, Hinrichs, W, Prester, A, Rohde, H, Aepfelbacher, M, Betzel, C. | | Deposit date: | 2021-10-17 | | Release date: | 2022-04-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural basis to repurpose boron-based proteasome inhibitors Bortezomib and Ixazomib as beta-lactamase inhibitors.
Sci Rep, 12, 2022
|
|
7SL1
 
 | |
7Q11
 
 | | Crystal structure of CTX-M-14 in complex with Ixazomib | | Descriptor: | Beta-lactamase, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Werner, N, Perbandt, M, Hinrichs, W, Prester, A, Rohde, H, Aepfelbacher, M, Betzel, C. | | Deposit date: | 2021-10-17 | | Release date: | 2022-04-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Structural basis to repurpose boron-based proteasome inhibitors Bortezomib and Ixazomib as beta-lactamase inhibitors.
Sci Rep, 12, 2022
|
|
8Q61
 
 | | Co-crystal structure of human AKT2 with compound 3 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-[4-(1-azanyl-3-methyl-3-oxidanyl-cyclobutyl)phenyl]-7-phenyl-1-propyl-pyrido[2,3-b][1,4]oxazin-2-one, GLYCEROL, ... | | Authors: | Harrison, T, Barker, O. | | Deposit date: | 2023-08-10 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Identification and development of a subtype-selective allosteric AKT inhibitor suitable for clinical development.
Sci Rep, 12, 2022
|
|
5NUC
 
 | | STAPHYLOCOCCAL NUCLEASE, 1-N-PENTANE THIOL DISULFIDE TO V23C VARIANT | | Descriptor: | CALCIUM ION, STAPHYLOCOCCAL NUCLEASE, THYMIDINE-3',5'-DIPHOSPHATE | | Authors: | Wynn, R, Harkins, P.C, Richards, F.M, Fox, R.O. | | Deposit date: | 1998-01-11 | | Release date: | 1998-04-29 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mobile unnatural amino acid side chains in the core of staphylococcal nuclease.
Protein Sci., 5, 1996
|
|
8BIR
 
 | | O-Methyltransferase Plu4895 in complex with SAH and AQ-256 | | Descriptor: | 1,3,8-tris(oxidanyl)anthracene-9,10-dione, CHLORIDE ION, IODIDE ION, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2022-11-02 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A set of closely related methyltransferases for site-specific tailoring of anthraquinone pigments.
Structure, 31, 2023
|
|
8WZO
 
 | | Parkin in complex with phospho NEDD8 | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase parkin, NEDD8, ... | | Authors: | Lenka, D.R, Kumar, A. | | Deposit date: | 2023-11-02 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Parkin in complex with phospho NEDD8
Structure
|
|
7SL7
 
 | |
8UVS
 
 | | Crystal structure of the wild-type Thermus thermophilus 70S ribosome in complex with spectinomycin derivative 2694, mRNA, deacylated A- and E-site tRNAphe, and deacylated P-site tRNAmet at 2.75A resolution | | Descriptor: | (2R,4R,4aS,5aR,6S,7S,8R,9S,9aR,10aS)-2-methyl-6,8-bis(methylamino)-4-({[2-(oxan-4-yl)ethyl]amino}methyl)octahydro-2H-pyrano[2,3-b][1,4]benzodioxine-4,4a,7,9(10aH)-tetrol, 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Killam, B.Y, Phelps, G.A, Lee, R.E, Polikanov, Y.S. | | Deposit date: | 2023-11-03 | | Release date: | 2024-08-07 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Development of 2nd generation aminomethyl spectinomycins that overcome native efflux in Mycobacterium abscessus.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
5AOH
 
 | | Crystal Structure of CarF | | Descriptor: | POTASSIUM ION, Spore coat protein CotH | | Authors: | Tichy, E.M, Hardwick, S.W, Luisi, B.F, C Salmond, G.P. | | Deposit date: | 2015-09-10 | | Release date: | 2017-01-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1.8 angstrom resolution crystal structure of the carbapenem intrinsic resistance protein CarF.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
8ZQU
 
 | | The crystal structure of PDE4D with isoaurostatin derivatives 2-6 | | Descriptor: | (3~{Z})-3-[[4-[bis(fluoranyl)methoxy]-3-cyclopentyloxy-phenyl]methylidene]-6-oxidanyl-1-benzofuran-2-one, 3',5'-cyclic-AMP phosphodiesterase 4D, MAGNESIUM ION, ... | | Authors: | Huang, Y.-Y, Luo, H.-B. | | Deposit date: | 2024-06-03 | | Release date: | 2025-02-12 | | Method: | X-RAY DIFFRACTION (2.00004554 Å) | | Cite: | Structure-based optimization of isoaurostatin as novel PDE4 inhibitors with anti-fibrotic effects
Chin.Chem.Lett., 2024
|
|
5OAD
 
 | | Crystal structure of mutant AChBP in complex with HEPES (T53F, Q74R, Y110A, I135S, G162E) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Dawson, A, Hunter, W.N, de Souza, J.O, Trumper, P. | | Deposit date: | 2017-06-21 | | Release date: | 2018-08-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Engineering a surrogate human heteromeric alpha / beta glycine receptor orthosteric site exploiting the structural homology and stability of acetylcholine-binding protein.
Iucrj, 6, 2019
|
|
8XFI
 
 | | High-resolution structure of the siderophore periplasmic binding protein FtsB from Streptococcus pyogenes with ferrioxamine E bound (crystal form 2) | | Descriptor: | (8E)-6,17,28-trihydroxy-1,6,12,17,23,28-hexaazacyclotritriacont-8-ene-2,5,13,16,24,27-hexone, CHLORIDE ION, FE (III) ION, ... | | Authors: | Caaveiro, J.M.M, Fernandez-Perez, J, Tsumoto, K. | | Deposit date: | 2023-12-13 | | Release date: | 2024-10-09 | | Last modified: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for the ligand promiscuity of the hydroxamate siderophore binding protein FtsB from Streptococcus pyogenes.
Structure, 32, 2024
|
|
5NU3
 
 | | Crystal structure of the human bromodomain of CREBBP bound to the inhibitor XDM-CBP | | Descriptor: | (R,R)-2,3-BUTANEDIOL, CREB-binding protein, SULFATE ION, ... | | Authors: | Huegle, M, Wohlwend, D. | | Deposit date: | 2017-04-28 | | Release date: | 2017-08-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Beyond the BET Family: Targeting CBP/p300 with 4-Acyl Pyrroles.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
8BJ2
 
 | | Crystal structure of Medicago truncatula histidinol-phosphate aminotransferase (HISN6) in the closed state | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETATE ION, SODIUM ION, ... | | Authors: | Rutkiewicz, M, Ruszkowski, M. | | Deposit date: | 2022-11-03 | | Release date: | 2023-03-22 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Insights into the substrate specificity, structure, and dynamics of plant histidinol-phosphate aminotransferase (HISN6).
Plant Physiol Biochem., 196, 2023
|
|
8UD6
 
 | | Crystal structure of the wild-type Thermus thermophilus 70S ribosome in complex with cresomycin, mRNA, deacylated A-site tRNAphe, aminoacylated P-site fMet-tRNAmet, and deacylated E-site tRNAphe at 2.70A resolution | | Descriptor: | (4S,5aS,8S,8aR)-4-(2-methylpropyl)-N-[(1R,5Z,7R,8R,9R,10R,11S,12R)-10,11,12-trihydroxy-7-methyl-13-oxa-2-thiabicyclo[7.3.1]tridec-5-en-8-yl]octahydro-2H-oxepino[2,3-c]pyrrole-8-carboxamide (non-preferred name), 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Aleksandrova, E.V, Syroegin, E.A, Wu, K.J.Y, Tresco, B.I.C, Ramkissoon, A, See, D.N.Y, Liow, P, Dittemore, G.A, Yu, M, Testolin, G, Mitcheltree, M.J, Liu, R.Y, Svetlov, M.S, Myers, A.G, Polikanov, Y.S. | | Deposit date: | 2023-09-28 | | Release date: | 2024-02-21 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | An antibiotic preorganized for ribosomal binding overcomes antimicrobial resistance.
Science, 383, 2024
|
|
8UD7
 
 | | Crystal structure of the A2058-N6-dimethylated Thermus thermophilus 70S ribosome in complex with cresomycin, mRNA, deacylated A-site tRNAphe, aminoacylated P-site fMet-tRNAmet, and deacylated E-site tRNAphe at 2.70A resolution | | Descriptor: | (4S,5aS,8S,8aR)-4-(2-methylpropyl)-N-[(1R,5Z,7R,8R,9R,10R,11S,12R)-10,11,12-trihydroxy-7-methyl-13-oxa-2-thiabicyclo[7.3.1]tridec-5-en-8-yl]octahydro-2H-oxepino[2,3-c]pyrrole-8-carboxamide (non-preferred name), 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Aleksandrova, E.V, Syroegin, E.A, Wu, K.J.Y, Tresco, B.I.C, Ramkissoon, A, See, D.N.Y, Liow, P, Dittemore, G.A, Yu, M, Testolin, G, Mitcheltree, M.J, Liu, R.Y, Svetlov, M.S, Myers, A.G, Polikanov, Y.S. | | Deposit date: | 2023-09-28 | | Release date: | 2024-02-21 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | An antibiotic preorganized for ribosomal binding overcomes antimicrobial resistance.
Science, 383, 2024
|
|
8UD8
 
 | | Crystal structure of the A2503-C2,C8-dimethylated Thermus thermophilus 70S ribosome in complex with cresomycin, mRNA, deacylated A-site tRNAphe, aminoacylated P-site fMet-tRNAmet, and deacylated E-site tRNAphe at 2.70A resolution | | Descriptor: | (4S,5aS,8S,8aR)-4-(2-methylpropyl)-N-[(1R,5Z,7R,8R,9R,10R,11S,12R)-10,11,12-trihydroxy-7-methyl-13-oxa-2-thiabicyclo[7.3.1]tridec-5-en-8-yl]octahydro-2H-oxepino[2,3-c]pyrrole-8-carboxamide (non-preferred name), 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Aleksandrova, E.V, Syroegin, E.A, Wu, K.J.Y, Tresco, B.I.C, Ramkissoon, A, See, D.N.Y, Liow, P, Dittemore, G.A, Yu, M, Testolin, G, Mitcheltree, M.J, Liu, R.Y, Svetlov, M.S, Myers, A.G, Polikanov, Y.S. | | Deposit date: | 2023-09-28 | | Release date: | 2024-02-21 | | Last modified: | 2025-02-12 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | An antibiotic preorganized for ribosomal binding overcomes antimicrobial resistance.
Science, 383, 2024
|
|
8ZQW
 
 | | The crystal structure of PDE4D with isoaurostatin derivatives 2-9 | | Descriptor: | (3~{Z})-3-[[4-[bis(fluoranyl)methoxy]-3-cyclopentyloxy-phenyl]methylidene]-6-oxidanyl-1~{H}-indol-2-one, 3',5'-cyclic-AMP phosphodiesterase 4D, MAGNESIUM ION, ... | | Authors: | Huang, Y.-Y, Luo, H.-B. | | Deposit date: | 2024-06-03 | | Release date: | 2025-02-12 | | Method: | X-RAY DIFFRACTION (2.100019 Å) | | Cite: | Structure-based optimization of isoaurostatin as novel PDE4 inhibitors with anti-fibrotic effects
Chin.Chem.Lett., 2024
|
|
8OWX
 
 | | Crystal Structure of METTL6 bound to SAH | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, S-ADENOSYL-L-HOMOCYSTEINE, tRNA N(3)-methylcytidine methyltransferase METTL6 | | Authors: | Throll, P, Basu, S, Dolce, L.G, Kowalinski, E. | | Deposit date: | 2023-04-28 | | Release date: | 2024-05-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Structural basis of tRNA recognition by the m 3 C RNA methyltransferase METTL6 in complex with SerRS seryl-tRNA synthetase.
Nat.Struct.Mol.Biol., 31, 2024
|
|
5O0V
 
 | |
