5CND
 
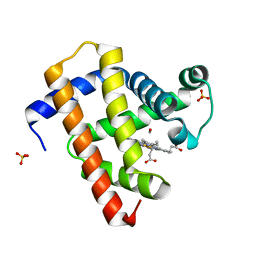 | | Ultrafast dynamics in myoglobin: 3 ps time delay | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Barends, T.R.M, Foucar, L, Ardevol, A, Nass, K.J, Aquila, A, Botha, S, Doak, R.B, Falahati, K, Hartmann, E, Hilpert, M, Heinz, M, Hoffmann, M.C, Koefinger, J, Koglin, J, Kovacsova, G, Liang, M, Milathianaki, D, Lemke, H.T, Reinstein, J, Roome, C.M, Shoeman, R.L, Williams, G.J, Burghardt, I, Hummer, G, Boutet, S, Schlichting, I. | | Deposit date: | 2015-07-17 | | Release date: | 2015-09-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Direct observation of ultrafast collective motions in CO myoglobin upon ligand dissociation.
Science, 350, 2015
|
|
4IDO
 
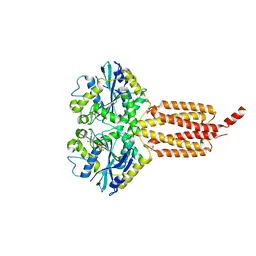 | | human atlastin-1 1-446, C-his6, GDPAlF4- | | Descriptor: | Atlastin-1, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Byrnes, L.J, Singh, A, Szeto, K, Benvin, N.M, O'Donnell, J.P, Zipfel, W.R, Sondermann, H. | | Deposit date: | 2012-12-12 | | Release date: | 2013-01-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.091 Å) | | Cite: | Structural basis for conformational switching and GTP loading of the large G protein atlastin.
Embo J., 32, 2013
|
|
4A81
 
 | | Crystal Structure of Major Birch Pollen Allergen Bet v 1 a in ternary complex with 8-Anilinonaphthalene-1-sulfonate (ANS) and deoxycholic acid | | Descriptor: | (3ALPHA,5BETA,12ALPHA)-3,12-DIHYDROXYCHOLAN-24-OIC ACID, (4S)-2-METHYL-2,4-PENTANEDIOL, 8-ANILINO-1-NAPHTHALENE SULFONATE, ... | | Authors: | Kofler, S, Brandstetter, H. | | Deposit date: | 2011-11-18 | | Release date: | 2012-05-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystallographically Mapped Ligand Binding Differs in High and Low Ige Binding Isoforms of Birch Pollen Allergen Bet V 1.
J.Mol.Biol., 422, 2012
|
|
4NFF
 
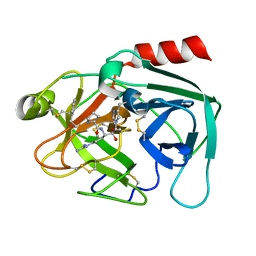 | | Human kallikrein-related peptidase 2 in complex with PPACK | | Descriptor: | D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, Kallikrein-2 | | Authors: | Skala, W, Brandstetter, H, Magdolen, V, Goettig, P. | | Deposit date: | 2013-10-31 | | Release date: | 2014-10-29 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-function analyses of human kallikrein-related peptidase 2 establish the 99-loop as master regulator of activity
J.Biol.Chem., 289, 2014
|
|
1ZGB
 
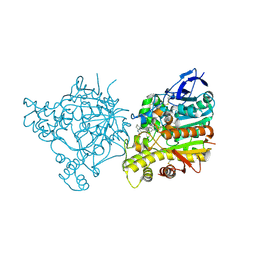 | | Crystal Structure of Torpedo Californica Acetylcholinesterase in Complex With an (R)-Tacrine(10)-Hupyridone Inhibitor. | | Descriptor: | (5R)-5-{[10-(1,2,3,4-TETRAHYDROACRIDIN-9-YLAMINO)DECYL]AMINO}-5,6,7,8-TETRAHYDROQUINOLIN-2(1H)-ONE, 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase | | Authors: | Haviv, H, Wong, D.M, Greenblatt, H.M, Carlier, P.R, Pang, Y.P, Silman, I, Sussman, J.L, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2005-04-21 | | Release date: | 2005-08-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Packing Mediates Enantioselective Ligand Recognition at the Peripheral Site of Acetylcholinesterase
J.Am.Chem.Soc., 127, 2005
|
|
4A87
 
 | |
5AN7
 
 | | Structure of the engineered retro-aldolase RA95.5-8F with a bound 1,3-diketone inhibitor | | Descriptor: | (2E)-1-(6-methoxynaphthalen-2-yl)but-2-en-1-one, PHOSPHATE ION, RA95.5-8F | | Authors: | Obexer, R, Mittl, P.R.E, Hilvert, D. | | Deposit date: | 2015-09-04 | | Release date: | 2016-08-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Emergence of a catalytic tetrad during evolution of a highly active artificial aldolase.
Nat Chem, 9, 2017
|
|
4A85
 
 | | Crystal Structure of Major Birch Pollen Allergen Bet v 1 a in complex with kinetin. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MAJOR POLLEN ALLERGEN BET V 1-A, ... | | Authors: | Kofler, S, Brandstetter, H. | | Deposit date: | 2011-11-18 | | Release date: | 2012-05-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystallographically Mapped Ligand Binding Differs in High and Low Ige Binding Isoforms of Birch Pollen Allergen Bet V 1.
J.Mol.Biol., 422, 2012
|
|
4N95
 
 | | E. coli sliding clamp in complex with 5-chloroindoline-2,3-dione | | Descriptor: | 5-chloro-1H-indole-2,3-dione, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Yin, Z, Oakley, A.J. | | Deposit date: | 2013-10-19 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of lead compounds targeting the bacterial sliding clamp using a fragment-based approach.
J.Med.Chem., 57, 2014
|
|
4A88
 
 | |
4A80
 
 | |
4AQO
 
 | |
4AR1
 
 | |
4AM4
 
 | | Bacterioferritin from Blastochloris viridis | | Descriptor: | BACTERIOFERRITIN, FE (III) ION, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Wahlgren, W.Y, Omran, H, von Stetten, D, Royant, A, van der Post, S, Katona, G. | | Deposit date: | 2012-03-07 | | Release date: | 2012-10-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural Characterization of Bacterioferritin from Blastochloris Viridis.
Plos One, 7, 2012
|
|
3AMG
 
 | | Crystal structures of Thermotoga maritima Cel5A in complex with Cellobiose substrate, mutant form | | Descriptor: | Endoglucanase, beta-D-glucopyranose, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Wu, T.H, Huang, C.H, Ko, T.P, Lai, H.L, Ma, Y, Cheng, Y.S, Liu, J.R, Guo, R.T. | | Deposit date: | 2010-08-20 | | Release date: | 2011-08-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Diverse substrate recognition mechanism revealed by Thermotoga maritima Cel5A structures in complex with cellotetraose, cellobiose and mannotriose
Biochim.Biophys.Acta, 1814, 2011
|
|
4ZWJ
 
 | | Crystal structure of rhodopsin bound to arrestin by femtosecond X-ray laser | | Descriptor: | Chimera protein of human Rhodopsin, mouse S-arrestin, and T4 Endolysin | | Authors: | Kang, Y, Zhou, X.E, Gao, X, He, Y, Liu, W, Ishchenko, A, Barty, A, White, T.A, Yefanov, O, Han, G.W, Xu, Q, de Waal, P.W, Ke, J, Tan, M.H.E, Zhang, C, Moeller, A, West, G.M, Pascal, B, Eps, N.V, Caro, L.N, Vishnivetskiy, S.A, Lee, R.J, Suino-Powell, K.M, Gu, X, Pal, K, Ma, J, Zhi, X, Boutet, S, Williams, G.J, Messerschmidt, M, Gati, C, Zatsepin, N.A, Wang, D, James, D, Basu, S, Roy-Chowdhury, S, Conrad, C, Coe, J, Liu, H, Lisova, S, Kupitz, C, Grotjohann, I, Fromme, R, Jiang, Y, Tan, M, Yang, H, Li, J, Wang, M, Zheng, Z, Li, D, Howe, N, Zhao, Y, Standfuss, J, Diederichs, K, Dong, Y, Potter, C.S, Carragher, B, Caffrey, M, Jiang, H, Chapman, H.N, Spence, J.C.H, Fromme, P, Weierstall, U, Ernst, O.P, Katritch, V, Gurevich, V.V, Griffin, P.R, Hubbell, W.L, Stevens, R.C, Cherezov, V, Melcher, K, Xu, H.E, GPCR Network (GPCR) | | Deposit date: | 2015-05-19 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.302 Å) | | Cite: | Crystal structure of rhodopsin bound to arrestin by femtosecond X-ray laser.
Nature, 523, 2015
|
|
4AM5
 
 | | Bacterioferritin from Blastochloris viridis | | Descriptor: | BACTERIOFERRITIN, FE (III) ION, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Wahlgren, W.Y, Omran, H, von Stetten, D, Royant, A, van der Post, S, Katona, G. | | Deposit date: | 2012-03-07 | | Release date: | 2012-10-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural Characterization of Bacterioferritin from Blastochloris Viridis.
Plos One, 7, 2012
|
|
3AOF
 
 | | Crystal structures of Thermotoga maritima Cel5A in complex with Mannotriose substrate | | Descriptor: | Endoglucanase, beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose | | Authors: | Wu, T.H, Huang, C.H, Ko, T.P, Lai, H.L, Ma, Y, Cheng, Y.S, Liu, J.R, Guo, R.T. | | Deposit date: | 2010-09-27 | | Release date: | 2011-08-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.288 Å) | | Cite: | Diverse substrate recognition mechanism revealed by Thermotoga maritima Cel5A structures in complex with cellotetraose, cellobiose and mannotriose
Biochim.Biophys.Acta, 1814, 2011
|
|
4JCF
 
 | |
4AW9
 
 | | Crystal structure of active legumain in complex with YVAD-CMK | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACE-TYR-VAL-ALA-ASP-CHLOROMETHYLKETONE, LEGUMAIN, ... | | Authors: | Dall, E, Brandstetter, H. | | Deposit date: | 2012-06-01 | | Release date: | 2013-06-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanistic and Structural Studies on Legumain Explain its Zymogenicity, Distinct Activation Pathways, and Regulation
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4B94
 
 | | Crystal structure of human Mps1 TPR domain | | Descriptor: | DI(HYDROXYETHYL)ETHER, DUAL SPECIFICITY PROTEIN KINASE TTK, GLYCEROL, ... | | Authors: | Littler, D, von Castelmur, E, De Marco, V, Perrakis, A. | | Deposit date: | 2012-08-31 | | Release date: | 2013-04-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Tpr Domain-Containing N-Terminal Module of Mps1 is Required for its Kinetochore Localization by Aurora B.
J.Cell Biol., 201, 2013
|
|
4NC3
 
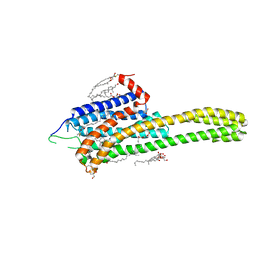 | | Crystal structure of the 5-HT2B receptor solved using serial femtosecond crystallography in lipidic cubic phase. | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHOLESTEROL, ... | | Authors: | Liu, W, Wacker, D, Gati, C, Han, G.W, James, D, Wang, D, Nelson, G, Weierstall, U, Katritch, V, Barty, A, Zatsepin, N.A, Li, D, Messerschmidt, M, Boutet, S, Williams, G.J, Koglin, J.E, Seibert, M.M, Wang, C, Shah, S.T.A, Basu, S, Fromme, R, Kupitz, C, Rendek, K.N, Grotjohann, I, Fromme, P, Kirian, R.A, Beyerlein, K.R, White, T.A, Chapman, H.N, Caffrey, M, Spence, J.C.H, Stevens, R.C, Cherezov, V, GPCR Network (GPCR) | | Deposit date: | 2013-10-23 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Serial femtosecond crystallography of G protein-coupled receptors.
Science, 342, 2013
|
|
1ZGC
 
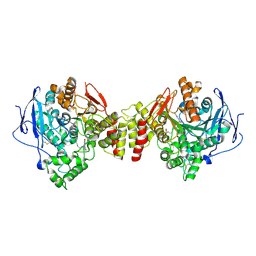 | | Crystal Structure of Torpedo Californica Acetylcholinesterase in Complex With an (RS)-Tacrine(10)-Hupyridone Inhibitor. | | Descriptor: | (5S)-5-{[10-(1,2,3,4-TETRAHYDROACRIDIN-9-YLAMINO)DECYL]AMINO}-5,6,7,8-TETRAHYDROQUINOLIN-2(1H)-ONE, 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase | | Authors: | Haviv, H, Wong, D.M, Greenblatt, H.M, Carlier, P.R, Pang, Y.P, Silman, I, Sussman, J.L, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2005-04-21 | | Release date: | 2005-08-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Packing Mediates Enantioselective Ligand Recognition at the Peripheral Site of Acetylcholinesterase
J.Am.Chem.Soc., 127, 2005
|
|
4B5Y
 
 | | X-ray structure of the cyan fluorescent protein mTurquoise-GL (K206A mutant) in space group C222(1) | | Descriptor: | GREEN FLUORESCENT PROTEIN | | Authors: | von Stetten, D, Lelimousin, M, Oost, K, Noirclerc-Savoye, M, Gadella, T.W.J, Goedhart, J, Royant, A. | | Deposit date: | 2012-08-08 | | Release date: | 2013-08-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Influence of the H148G Mutation on Fluorescence Properties of Cyan Fluorescent Proteins
To be Published
|
|
4AS8
 
 | | X-ray structure of the cyan fluorescent protein Cerulean cryoprotected with ethylene glycol | | Descriptor: | 1,2-ETHANEDIOL, GREEN FLUORESCENT PROTEIN, MAGNESIUM ION | | Authors: | von Stetten, D, Batot, G, Noirclerc-Savoye, M, Royant, A. | | Deposit date: | 2012-04-29 | | Release date: | 2012-10-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Alteration of Fluorescent Protein Spectroscopic Properties Upon Cryoprotection
Acta Crystallogr.,Sect.D, 68, 2012
|
|
