7QHW
 
 | | TTBK1 kinase domain in complex with inhibitor 29 | | Descriptor: | GLYCEROL, SULFATE ION, Tau-tubulin kinase 1, ... | | Authors: | Nozal, V, Liehta, D. | | Deposit date: | 2021-12-14 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | TDP-43 Modulation by Tau-Tubulin Kinase 1 Inhibitors: A New Avenue for Future Amyotrophic Lateral Sclerosis Therapy.
J.Med.Chem., 65, 2022
|
|
7QKR
 
 | | Cryo-EM structure of ABC transporter STE6-2p from Pichia pastoris with Verapamil at 3.2 A resolution | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, Dexverapamil, MAGNESIUM ION, ... | | Authors: | Schleker, E.S.M, Reinhart, C. | | Deposit date: | 2021-12-18 | | Release date: | 2022-10-26 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural and functional investigation of ABC transporter STE6-2p from Pichia pastoris reveals unexpected interaction with sterol molecules.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7CHY
 
 | |
7R7R
 
 | | Structure of Human Anaplastic Lymphoma Kinase Domain in complex with ((2~{R})-2-[5-[6-amino-5-[(1~{R})-1-[5-fluoro-2-(triazol-2-yl)phenyl]ethoxy]-3-pyridyl]-4-methyl-thiazol-2-yl]propane-1,2-diol) | | Descriptor: | (2R)-2-[5-(6-amino-5-{(1R)-1-[2-(1,3-dihydro-2H-1,2,3-triazol-2-yl)-5-fluorophenyl]ethoxy}pyridin-3-yl)-4-methyl-1,3-thiazol-2-yl]propane-1,2-diol, ALK tyrosine kinase receptor | | Authors: | McTigue, M. | | Deposit date: | 2021-06-25 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.935 Å) | | Cite: | Analysis of lorlatinib analogs reveals a roadmap for targeting diverse compound resistance mutations in ALK-positive lung cancer.
Nat Cancer, 3, 2022
|
|
7R7K
 
 | |
7RAR
 
 | |
7RJ2
 
 | |
7RJ4
 
 | |
7CHZ
 
 | |
7RHN
 
 | |
7RHM
 
 | |
7RMJ
 
 | | Disulfide stabilized HIV-1 CA hexamer in complex with capsid inhibitor (S)-N-(1-(3-(4-chloro-3-(methylsulfonamido)-1-(2,2,2-trifluoroethyl)-1H-indazol-7-yl)-6-(3-methyl-3-(methylsulfonyl)but-1-yn-1-yl)pyridin-2-yl)-2-(3,5-difluorophenyl)ethyl)-2-(3-(trifluoromethyl)-4,5,6,7-tetrahydro-1H-indazol-1-yl)acetamide | | Descriptor: | CAPSID PROTEIN P24, CHLORIDE ION, IODIDE ION, ... | | Authors: | Bester, S.M, Kvaratskhelia, M. | | Deposit date: | 2021-07-27 | | Release date: | 2022-08-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structural and Mechanistic Bases of Viral Resistance to HIV-1 Capsid Inhibitor Lenacapavir.
Mbio, 13, 2022
|
|
7RMM
 
 | |
8CCE
 
 | | The Fk1 domain of FKBP51 in complex with 2-(3-((R)-1-(((S)-1-((S)-2-(5-chlorothiophen-2-yl)butanoyl)piperidine-2-carbonyl)oxy)-3-(3,4-dimethoxyphenyl)propyl)phenoxy)acetic acid | | Descriptor: | 2-[3-[(1~{R})-1-[(2~{S})-1-[(2~{S})-2-(5-chloranylthiophen-2-yl)butanoyl]piperidin-2-yl]carbonyloxy-3-(3,4-dimethoxyphenyl)propyl]phenoxy]ethanoic acid, GLYCEROL, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Knaup, F.H, Walz, C.M, Hausch, F. | | Deposit date: | 2023-01-27 | | Release date: | 2023-04-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-Based Discovery of a New Selectivity-Enabling Motif for the FK506-Binding Protein 51.
J.Med.Chem., 66, 2023
|
|
8CCC
 
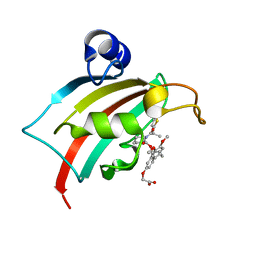 | | The Fk1 domain of FKBP51 in complex with 2-(3-((R)-1-(((S)-1-((S)-2-(5-chlorothiophen-2-yl)propanoyl)piperidine-2-carbonyl)oxy)-3-(3,4-dimethoxyphenyl)propyl)phenoxy)acetic acid | | Descriptor: | 2-[3-[(1~{R})-1-[(2~{S})-1-[(2~{S})-2-(5-chloranylthiophen-2-yl)propanoyl]piperidin-2-yl]carbonyloxy-3-(3,4-dimethoxyphenyl)propyl]phenoxy]ethanoic acid, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Knaup, F.H, Walz, C.M, Hausch, F. | | Deposit date: | 2023-01-27 | | Release date: | 2023-04-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure-Based Discovery of a New Selectivity-Enabling Motif for the FK506-Binding Protein 51.
J.Med.Chem., 66, 2023
|
|
4PIS
 
 | | Crystal structure of human adenovirus 8 protease in complex with a nitrile inhibitor | | Descriptor: | N~2~-[(2R)-2-(3,5-dichlorophenyl)-2-(dimethylamino)acetyl]-N-({2-[(Z)-iminomethyl]pyrimidin-4-yl}methyl)-L-isoleucinamide, PVI, Protease | | Authors: | Mac Sweeney, A, Grosche, P, Ellis, D, Combrink, K, Erbel, P, Hughes, N, Sirockin, F, Melkko, S, Bernardi, A, Ramage, P, Jarousse, N, Altmann, E. | | Deposit date: | 2014-05-09 | | Release date: | 2014-09-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery and structure-based optimization of adenain inhibitors.
Acs Med.Chem.Lett., 5, 2014
|
|
4PIV
 
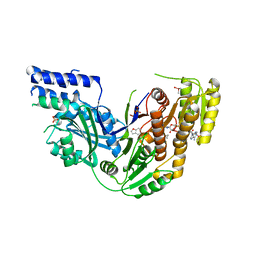 | | Human Fatty Acid Synthase Psi/KR Tri-Domain with NADPH and GSK2194069 | | Descriptor: | 4-[4-(1-benzofuran-5-yl)phenyl]-5-{[(3S)-1-(cyclopropylcarbonyl)pyrrolidin-3-yl]methyl}-2,4-dihydro-3H-1,2,4-triazol-3-one, CACODYLATE ION, Fatty acid synthase, ... | | Authors: | Williams, S.P, Wang, L, Brown, K.K, Parrish, C.A, Hardwicke, M.A. | | Deposit date: | 2014-05-09 | | Release date: | 2014-07-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | A human fatty acid synthase inhibitor binds beta-ketoacyl reductase in the keto-substrate site.
Nat.Chem.Biol., 10, 2014
|
|
6XB2
 
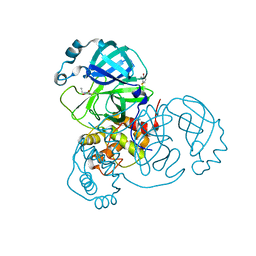 | | Room temperature X-ray crystallography reveals catalytic cysteine in the SARS-CoV-2 3CL Mpro is highly reactive: Insights for enzyme mechanism and drug design | | Descriptor: | 1-ETHYL-PYRROLIDINE-2,5-DIONE, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Kneller, D.W, Kovalevsky, A, Coates, L. | | Deposit date: | 2020-06-05 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Room-temperature X-ray crystallography reveals the oxidation and reactivity of cysteine residues in SARS-CoV-2 3CL M pro : insights into enzyme mechanism and drug design.
Iucrj, 7, 2020
|
|
6DUK
 
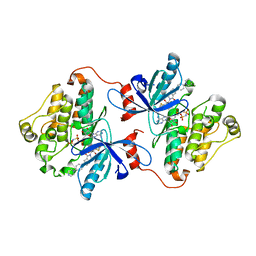 | | EGFR with an allosteric inhibitor | | Descriptor: | (2R)-2-(5-fluoro-2-hydroxyphenyl)-2-{1-oxo-6-[4-(piperazin-1-yl)phenyl]-1,3-dihydro-2H-isoindol-2-yl}-N-(1,3-thiazol-2-yl)acetamide, Epidermal growth factor receptor, MAGNESIUM ION, ... | | Authors: | Park, E, Eck, M.J. | | Deposit date: | 2018-06-21 | | Release date: | 2019-06-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Single and Dual Targeting of Mutant EGFR with an Allosteric Inhibitor.
Cancer Discov, 9, 2019
|
|
8C25
 
 | | purine nucleoside phosphorylase in complex with JS-375 | | Descriptor: | CHLORIDE ION, GLYCEROL, Purine nucleoside phosphorylase, ... | | Authors: | Djukic, S, Pachl, P, Rezacova, P. | | Deposit date: | 2022-12-21 | | Release date: | 2023-05-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Design, Synthesis, Biological Evaluation, and Crystallographic Study of Novel Purine Nucleoside Phosphorylase Inhibitors.
J.Med.Chem., 66, 2023
|
|
4PF9
 
 | | Crystal structure of insulin degrading enzyme complexed with inhibitor | | Descriptor: | Insulin-degrading enzyme, ZINC ION, methyl [(2S)-2-[4-({5-[4-({(2S)-2-[(3S)-3-amino-2-oxopiperidin-1-yl]-2-cyclohexylacetyl}amino)phenyl]pentyl}oxy)phenyl]-3-(quinolin-3-yl)propyl]carbamate | | Authors: | Wang, Y, Guo, S. | | Deposit date: | 2014-04-28 | | Release date: | 2015-06-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Dual Exosite-binding Inhibitors of Insulin-degrading Enzyme Challenge Its Role as the Primary Mediator of Insulin Clearance in Vivo.
J.Biol.Chem., 290, 2015
|
|
7ZFX
 
 | | VDR complex with Aromatic-D-Ring Analog | | Descriptor: | (1R,3S,5Z)-5-[(E)-3-[3,5-bis(6-methyl-6-oxidanyl-heptyl)phenyl]prop-2-enylidene]-4-methylidene-cyclohexane-1,3-diol, ACETATE ION, Nuclear receptor coactivator 1, ... | | Authors: | Rochel, N. | | Deposit date: | 2022-04-01 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Design, Synthesis, Biological Activity, and Structural Analysis of Novel Des-C-Ring and Aromatic-D-Ring Analogues of 1 alpha ,25-Dihydroxyvitamin D 3.
J.Med.Chem., 65, 2022
|
|
4PUB
 
 | | Crystal structure of Fab DX-2930 | | Descriptor: | CHLORIDE ION, DX-2930 HEAVY CHAIN, DX-2930 LIGHT CHAIN | | Authors: | Abendroth, J, Edwards, T.E, Nixon, A, Ladner, R. | | Deposit date: | 2014-03-12 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Inhibition of plasma kallikrein by a highly specific active site blocking antibody.
J.Biol.Chem., 289, 2014
|
|
6DK0
 
 | | Human sigma-1 receptor bound to NE-100 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, GLYCEROL, N-{2-[4-methoxy-3-(2-phenylethoxy)phenyl]ethyl}-N-propylpropan-1-amine, ... | | Authors: | Schmidt, H.R, Kruse, A.C. | | Deposit date: | 2018-05-28 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for sigma1receptor ligand recognition.
Nat. Struct. Mol. Biol., 25, 2018
|
|
4ZX5
 
 | |
