2L0T
 
 | | Solution structure of the complex of ubiquitin and the VHS domain of Stam2 | | Descriptor: | Signal transducing adapter molecule 2, Ubiquitin | | Authors: | Lange, A, Hoeller, D, Wienk, H, Marcillat, O, Lancelin, J, Walker, O. | | Deposit date: | 2010-07-15 | | Release date: | 2010-12-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR reveals a different mode of binding of the Stam2 VHS domain to ubiquitin and diubiquitin.
Biochemistry, 50, 2011
|
|
5NKZ
 
 | | Crystal structure of H. polymorpha ubiquitin conjugating enzyme Pex4p in complex with soluble domain of Pex22p | | Descriptor: | Peroxin 22, Ubiquitin-conjugating enzyme E2-21 kDa | | Authors: | Danda, N, Lunev, S, Ali, A, Groves, M.R, Williams, C. | | Deposit date: | 2017-04-03 | | Release date: | 2018-01-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural insights into K48-linked ubiquitin chain formation by the Pex4p-Pex22p complex.
Biochem. Biophys. Res. Commun., 496, 2018
|
|
2JT4
 
 | |
2EKE
 
 | |
4M0W
 
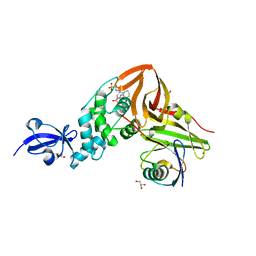 | | Crystal Structure of SARS-CoV papain-like protease C112S mutant in complex with ubiquitin | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, GLYCEROL, Replicase polyprotein 1a, ... | | Authors: | Chou, C.-Y, Chen, H.-Y, Lai, H.-Y, Cheng, S.-C, Chou, Y.-W. | | Deposit date: | 2013-08-02 | | Release date: | 2014-02-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis for catalysis and ubiquitin recognition by the severe acute respiratory syndrome coronavirus papain-like protease
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4AE4
 
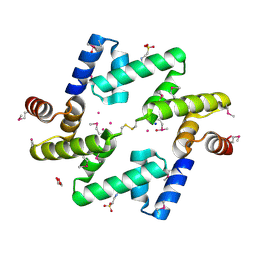 | | The UBAP1 subunit of ESCRT-I interacts with ubiquitin via a novel SOUBA domain | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, GLYCEROL, POTASSIUM ION, ... | | Authors: | Agromayor, M, Soler, N, Caballe, A, Kueck, T, Freund, S.M, Allen, M.D, Bycroft, M, Perisic, O, Ye, Y, McDonald, B, Scheel, H, Hofmann, K, Neil, S.J.D, Martin-Serrano, J, Williams, R.L. | | Deposit date: | 2012-01-06 | | Release date: | 2012-03-21 | | Last modified: | 2018-02-28 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The UBAP1 subunit of ESCRT-I interacts with ubiquitin via a SOUBA domain.
Structure, 20, 2012
|
|
2ETL
 
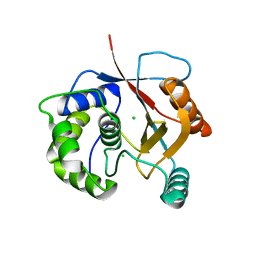 | | Crystal Structure of Ubiquitin Carboxy-terminal Hydrolase L1 (UCH-L1) | | Descriptor: | CHLORIDE ION, Ubiquitin carboxyl-terminal hydrolase isozyme L1 | | Authors: | Das, C, Hoang, Q.Q, Kreinbring, C.A, Luchansky, S.J, Meray, R.K, Ray, S.S, Lansbury, P.T, Ringe, D, Petsko, G.A. | | Deposit date: | 2005-10-27 | | Release date: | 2006-03-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for conformational plasticity of the Parkinson's disease-associated ubiquitin hydrolase UCH-L1.
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
2MBH
 
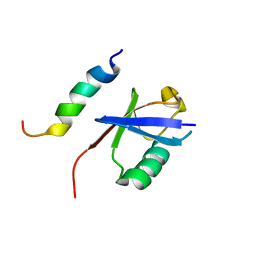 | | NMR structure of EKLF(22-40)/Ubiquitin Complex | | Descriptor: | Krueppel-like factor 1, Ubiquitin | | Authors: | Raiola, L, Omichinski, J.G. | | Deposit date: | 2013-07-31 | | Release date: | 2013-10-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Characterization of a Noncovalent Complex between Ubiquitin and the Transactivation Domain of the Erythroid-Specific Factor EKLF.
Structure, 21, 2013
|
|
6NFT
 
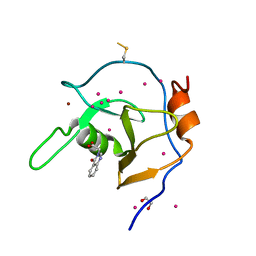 | | Structure of USP5 zinc-finger ubiquitin binding domain co-crystallized with (4-oxoquinazolin-3(4H)-yl)acetic acid | | Descriptor: | (4-oxoquinazolin-3(4H)-yl)acetic acid, 1,2-ETHANEDIOL, UNKNOWN ATOM OR ION, ... | | Authors: | Harding, R.J, Mann, M.K, Tempel, W, Bountra, C, Arrowmsmith, C.M, Edwards, A.M, Schapira, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-12-20 | | Release date: | 2019-01-02 | | Last modified: | 2021-06-09 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Discovery of Small Molecule Antagonists of the USP5 Zinc Finger Ubiquitin-Binding Domain.
J.Med.Chem., 62, 2019
|
|
2HD5
 
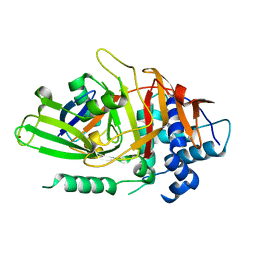 | | USP2 in complex with ubiquitin | | Descriptor: | Polyubiquitin, Ubiquitin carboxyl-terminal hydrolase 2, ZINC ION | | Authors: | Renatus, M, Kroemer, M. | | Deposit date: | 2006-06-20 | | Release date: | 2006-08-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Basis of Ubiquitin Recognition by the Deubiquitinating Protease USP2.
Structure, 14, 2006
|
|
2YLM
 
 | |
5O76
 
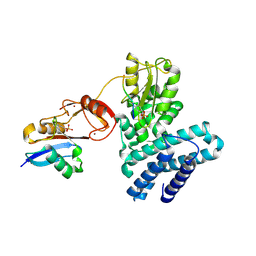 | | Structure of phosphoY371 c-CBL in complex with ZAP70-peptide and UbV.pCBL ubiquitin variant | | Descriptor: | CALCIUM ION, E3 ubiquitin-protein ligase CBL, Tyrosine protein kinase ZAP70 peptide, ... | | Authors: | Gabrielsen, M, Buetow, L, Huang, D.T. | | Deposit date: | 2017-06-08 | | Release date: | 2017-11-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.473 Å) | | Cite: | A General Strategy for Discovery of Inhibitors and Activators of RING and U-box E3 Ligases with Ubiquitin Variants.
Mol. Cell, 68, 2017
|
|
2OOA
 
 | |
4RF1
 
 | | Crystal structure of the Middle-East respiratory syndrome coronavirus papain-like protease in complex with ubiquitin (space group P63) | | Descriptor: | 3-AMINOPROPANE, ORF1ab protein, S-1,2-PROPANEDIOL, ... | | Authors: | Bailey-Elkin, B.A, Johnson, G.G, Mark, B.L. | | Deposit date: | 2014-09-24 | | Release date: | 2014-10-22 | | Last modified: | 2015-01-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structure of the Middle East Respiratory Syndrome Coronavirus (MERS-CoV) Papain-like Protease Bound to Ubiquitin Facilitates Targeted Disruption of Deubiquitinating Activity to Demonstrate Its Role in Innate Immune Suppression.
J.Biol.Chem., 289, 2014
|
|
4RF0
 
 | | Crystal structure of the Middle-East respiratory syndrome coronavirus papain-like protease in complex with ubiquitin (space group P6522) | | Descriptor: | 3-AMINOPROPANE, ORF1ab protein, SULFATE ION, ... | | Authors: | Bailey-Elkin, B.A, Johnson, G.G, Mark, B.L. | | Deposit date: | 2014-09-24 | | Release date: | 2014-10-22 | | Last modified: | 2015-01-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of the Middle East Respiratory Syndrome Coronavirus (MERS-CoV) Papain-like Protease Bound to Ubiquitin Facilitates Targeted Disruption of Deubiquitinating Activity to Demonstrate Its Role in Innate Immune Suppression.
J.Biol.Chem., 289, 2014
|
|
2JY6
 
 | |
6NXL
 
 | | Ubiquitin binding variants | | Descriptor: | Polyubiquitin-B | | Authors: | Miller, D.J, Watson, E.R. | | Deposit date: | 2019-02-08 | | Release date: | 2020-01-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.803 Å) | | Cite: | Protein engineering of a ubiquitin-variant inhibitor of APC/C identifies a cryptic K48 ubiquitin chain binding site.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6NJD
 
 | |
5OJJ
 
 | | Crystal structure of the Zn-bound ubiquitin-conjugating enzyme Ube2T | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, Ubiquitin-conjugating enzyme E2 T, ... | | Authors: | Morreale, F.E, Testa, A, Chaugule, V.K, Bortoluzzi, A, Ciulli, A, Walden, H. | | Deposit date: | 2017-07-21 | | Release date: | 2017-10-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Mind the Metal: A Fragment Library-Derived Zinc Impurity Binds the E2 Ubiquitin-Conjugating Enzyme Ube2T and Induces Structural Rearrangements.
J. Med. Chem., 60, 2017
|
|
2LXH
 
 | | NMR structure of the RING domain in ubiquitin ligase gp78 | | Descriptor: | E3 ubiquitin-protein ligase AMFR, ZINC ION | | Authors: | Das, R, Linag, Y, Mariano, J, Li, J, Huang, T, King, A, Weissman, A, Ji, X, Byrd, R. | | Deposit date: | 2012-08-27 | | Release date: | 2013-08-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Allosteric regulation of E2:E3 interactions promote a processive ubiquitination machine.
Embo J., 32, 2013
|
|
5DKA
 
 | | A C2HC zinc finger is essential for the activity of the RING ubiquitin ligase RNF125 | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase RNF125, MAGNESIUM ION, ... | | Authors: | Boer, D.R, Coll, M, Bijlmakers, M.J. | | Deposit date: | 2015-09-03 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A C2HC zinc finger is essential for the RING-E2 interaction of the ubiquitin ligase RNF125.
Sci Rep, 6, 2016
|
|
6JMA
 
 | | cryo-EM structure of DOT1L bound to H2B ubiquitinated nucleosome | | Descriptor: | DNA I&J, Histone H2A, Histone H2B 1.1, ... | | Authors: | Jang, S, Song, J.J. | | Deposit date: | 2019-03-07 | | Release date: | 2019-05-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Structural basis of recognition and destabilization of the histone H2B ubiquitinated nucleosome by the DOT1L histone H3 Lys79 methyltransferase.
Genes Dev., 33, 2019
|
|
5LP8
 
 | |
2GBM
 
 | | Crystal Structure of the 35-36 8 Glycine Insertion Mutant of Ubiquitin | | Descriptor: | ARSENIC, Ubiquitin | | Authors: | Ferraro, D.M, Ferraro, D.J, Ramaswamy, S, Robertson, A.D. | | Deposit date: | 2006-03-10 | | Release date: | 2006-05-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structures of Ubiquitin Insertion Mutants Support Site-specific Reflex Response to Insertions Hypothesis.
J.Mol.Biol., 359, 2006
|
|
2NSQ
 
 | | Crystal structure of the C2 domain of the human E3 ubiquitin-protein ligase NEDD4-like protein | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase NEDD4-like protein, GLYCEROL | | Authors: | Walker, J.R, Avvakumov, G.V, Xue, S, Butler-Cole, C, Finerty Jr, P.J, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-11-06 | | Release date: | 2006-12-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The C2 domain of the human E3 ubiquitin-protein ligase NEDD4-like protein
To be Published
|
|
