7V2A
 
 | | SARS-CoV-2 Spike trimer in complex with XG014 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, The heavy chain of XG014, ... | | Authors: | Wang, K, Wang, X, Pan, L. | | Deposit date: | 2021-08-07 | | Release date: | 2021-10-20 | | Last modified: | 2022-07-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | An ultrapotent pan-beta-coronavirus lineage B ( beta-CoV-B) neutralizing antibody locks the receptor-binding domain in closed conformation by targeting its conserved epitope.
Protein Cell, 13, 2022
|
|
2GSG
 
 | | Crystal structure of the Fv fragment of a monoclonal antibody specific for poly-glutamine | | Descriptor: | SULFATE ION, monoclonal antibody heavy chain, monoclonal antibody light chain | | Authors: | Li, P, Huey-Tubman, K.E, West Jr, A.P, Bennett, M.J, Bjorkman, P.J. | | Deposit date: | 2006-04-26 | | Release date: | 2007-04-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure of a polyQ-anti-polyQ complex reveals binding according to a linear lattice model.
Nat.Struct.Mol.Biol., 14, 2007
|
|
8BSE
 
 | | CRYSTAL STRUCTURE OF SARS-COV-2 RECEPTOR BINDING DOMAIN (RBD) in complex with 1D1 Fab | | Descriptor: | 1D1 FAB HEAVY CHAIN, 1D1 FAB LIGHT CHAIN, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Welin, M, Kimbung, Y.R, Focht, D, Pisitkun, T. | | Deposit date: | 2022-11-25 | | Release date: | 2023-05-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Efficacy of the combination of monoclonal antibodies against the SARS-CoV-2 Beta and Delta variants.
Plos One, 18, 2023
|
|
8BSF
 
 | | CRYSTAL STRUCTURE OF SARS-COV-2 RECEPTOR BINDING DOMAIN (RBD-beta variant) in complex with 3D2 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3D2 FAB HEAVY CHAIN, 3D2 FAB LIGHT CHAIN, ... | | Authors: | Welin, M, Kimbung, Y.R, Focht, D, Pisitkun, T. | | Deposit date: | 2022-11-25 | | Release date: | 2023-05-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Efficacy of the combination of monoclonal antibodies against the SARS-CoV-2 Beta and Delta variants.
Plos One, 18, 2023
|
|
7TYV
 
 | | Structure of Lassa Virus glycoprotein (Josiah) bound to Fab 25.10C | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 25.10C Fab Heavy Chain, ... | | Authors: | Enriquez, A.S, Hastie, K.M. | | Deposit date: | 2022-02-14 | | Release date: | 2022-06-01 | | Last modified: | 2022-06-15 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Delineating the mechanism of anti-Lassa virus GPC-A neutralizing antibodies.
Cell Rep, 39, 2022
|
|
7RU1
 
 | | SARS-CoV-2-6P-Mut7 S protein (C3 symmetry) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Ozorowski, G, Turner, H.L, Ward, A.B. | | Deposit date: | 2021-08-16 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Engineering SARS-CoV-2 neutralizing antibodies for increased potency and reduced viral escape pathways.
Iscience, 25, 2022
|
|
7RU2
 
 | | SARS-CoV-2-6P-Mut7 S protein (asymmetric) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Ozorowski, G, Turner, H.L, Ward, A.B. | | Deposit date: | 2021-08-16 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Engineering SARS-CoV-2 neutralizing antibodies for increased potency and reduced viral escape pathways.
Iscience, 25, 2022
|
|
7K9K
 
 | |
7K9H
 
 | | SARS-CoV-2 Spike in complex with neutralizing Fab 2B04 (one up, two down conformation) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2B04 heavy chain, ... | | Authors: | Errico, J.M, Fremont, D.H, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-09-29 | | Release date: | 2021-09-29 | | Last modified: | 2021-11-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural mechanism of SARS-CoV-2 neutralization by two murine antibodies targeting the RBD.
Cell Rep, 37, 2021
|
|
2HWZ
 
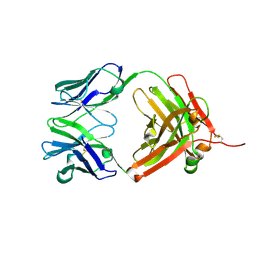 | | Fab fragment of Humanized anti-viral antibody MEDI-493 (Synagis TM) | | Descriptor: | Immunoglobulin Fab heavy chain, Immunoglobulin Fab light chain | | Authors: | Braden, B. | | Deposit date: | 2006-08-02 | | Release date: | 2007-08-14 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification of a single tryptophan residue as critical for binding activity in a humanized monoclonal antibody against respiratory syncytial virus
To be Published
|
|
3KDM
 
 | |
7LJR
 
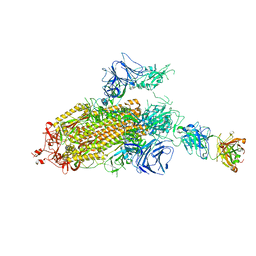 | | SARS-CoV-2 Spike Protein Trimer bound to DH1043 fab | | Descriptor: | Fab DH1043 heavy chain, Fab DH1043 light chain, Spike glycoprotein | | Authors: | Gobeil, S, Acharya, P. | | Deposit date: | 2021-01-30 | | Release date: | 2021-03-31 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | The functions of SARS-CoV-2 neutralizing and infection-enhancing antibodies in vitro and in mice and nonhuman primates.
Biorxiv, 2021
|
|
8AWM
 
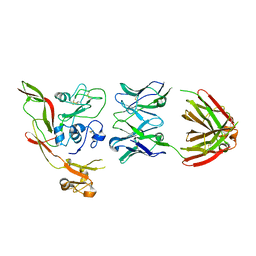 | | RVFV GnH with Fab268 bound | | Descriptor: | Glycoprotein, Heavy chain Fab268, Light chain Fab268 | | Authors: | Hulswit, R.J.G, Bowden, T.A, Stass, R. | | Deposit date: | 2022-08-30 | | Release date: | 2023-09-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Multifunctional human monoclonal antibody combination mediates protection against Rift Valley fever virus at low doses.
Nat Commun, 14, 2023
|
|
5NPJ
 
 | |
7A3N
 
 | | Crystal structure of Zika virus envelope glycoprotein in complex with the Fab fragment of the broadly neutralizing human antibody EDE1 C10 | | Descriptor: | CALCIUM ION, Core protein, EDE1 C10 Fab | | Authors: | Sharma, A, Vaney, M.C, Guardado-Calvo, P, Duquerroy, S, Rouvinski, A, Rey, F.A. | | Deposit date: | 2020-08-18 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The epitope arrangement on flavivirus particles contributes to Mab C10's extraordinary neutralization breadth across Zika and dengue viruses.
Cell, 184, 2021
|
|
7S8H
 
 | |
2VIS
 
 | | INFLUENZA VIRUS HEMAGGLUTININ, (ESCAPE) MUTANT WITH THR 131 REPLACED BY ILE, COMPLEXED WITH A NEUTRALIZING ANTIBODY | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HEMAGGLUTININ, IMMUNOGLOBULIN (IGG1, ... | | Authors: | Bizebard, T, Fleury, D, Gigant, B, Wharton, S.A, Skehel, J.J, Knossow, M. | | Deposit date: | 1997-12-22 | | Release date: | 1998-04-29 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Antigen distortion allows influenza virus to escape neutralization.
Nat.Struct.Biol., 5, 1998
|
|
2VIT
 
 | | INFLUENZA VIRUS HEMAGGLUTININ, MUTANT WITH THR 155 REPLACED BY ILE, COMPLEXED WITH A NEUTRALIZING ANTIBODY | | Descriptor: | HEMAGGLUTININ, IMMUNOGLOBULIN (IGG1, LAMBDA), ... | | Authors: | Bizebard, T, Fleury, D, Gigant, B, Wharton, S.A, Skehel, J.J, Knossow, M. | | Deposit date: | 1997-12-22 | | Release date: | 1998-04-29 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Antigen distortion allows influenza virus to escape neutralization.
Nat.Struct.Biol., 5, 1998
|
|
3LES
 
 | | 2F5 Epitope scaffold ES2 | | Descriptor: | RNA polymerase sigma factor, SULFATE ION | | Authors: | Ofek, G, Guenaga, F.J, Schief, W.R, Skinner, J, Wyatt, R, Baker, D, Kwong, P.D. | | Deposit date: | 2010-01-15 | | Release date: | 2010-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Elicitation of structure-specific antibodies by epitope scaffolds.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
8T4K
 
 | | MD64 N332-GT5 SOSIP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, MD64 N332-GT5 SOSIP gp120, ... | | Authors: | Ozorowski, G, Torres, J.L, Ward, A.B. | | Deposit date: | 2023-06-09 | | Release date: | 2024-05-08 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Vaccine priming of rare HIV broadly neutralizing antibody precursors in nonhuman primates.
Science, 384, 2024
|
|
7V26
 
 | | XG005-bound SARS-CoV-2 S | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, XG005 Heavy chain, ... | | Authors: | Zhan, W.Q, Zhang, X, Sun, L, Chen, Z.G. | | Deposit date: | 2021-08-07 | | Release date: | 2021-10-20 | | Last modified: | 2022-07-06 | | Method: | ELECTRON MICROSCOPY (3.85 Å) | | Cite: | An ultrapotent pan-beta-coronavirus lineage B ( beta-CoV-B) neutralizing antibody locks the receptor-binding domain in closed conformation by targeting its conserved epitope.
Protein Cell, 13, 2022
|
|
1RIU
 
 | | Anti-Cocaine Antibody M82G2 Complexed With Norbenzoylecgonine | | Descriptor: | 3-(BENZOYLOXY)-8-AZA-BICYCLO[3.2.1]OCTANE-2-CARBOXYLIC ACID, Fab M82G2, Heavy Chain, ... | | Authors: | Pozharski, E, Hewagama, A, Shanafelt, A, Petsko, G, Ringe, D. | | Deposit date: | 2003-11-18 | | Release date: | 2003-12-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Carving a Binding Site: Structural Study of an Anti-Cocaine Antibody in Complex with Three Cocaine Analogs
To be Published
|
|
7M8K
 
 | | Cryo-EM structure of Brazil (P.1) SARS-CoV-2 spike glycoprotein variant in the prefusion state (1 RBD up) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Casner, R.G, Cerutti, G, Shapiro, L, Ho, D.D. | | Deposit date: | 2021-03-29 | | Release date: | 2021-05-05 | | Last modified: | 2021-05-26 | | Method: | ELECTRON MICROSCOPY | | Cite: | Increased resistance of SARS-CoV-2 variant P.1 to antibody neutralization.
Cell Host Microbe, 29, 2021
|
|
1RIV
 
 | | Anti-Cocaine Antibody M82G2 Complexed With meta-Oxybenzoylecgonine | | Descriptor: | 3-(3-HYDROXY-BENZOYLOXY)-8-METHYL-8-AZA-BICYCLO[3.2.1]OCTANE-2-CARBOXYLIC ACID, Fab M82G2, Heavy Chain, ... | | Authors: | Pozharski, E, Hewagama, A, Shanafelt, A, Petsko, G, Ringe, D. | | Deposit date: | 2003-11-18 | | Release date: | 2003-12-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Carving a Binding Site: Structural Study of an Anti-Cocaine Antibody in Complex with Three Cocaine Analogs
To be Published
|
|
4WV1
 
 | |
